6WTH
 
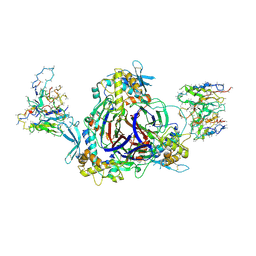 | | Full-length human ENaC ECD | | 分子名称: | 10D4 Fab, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Posert, R, Baconguis, I, Noreng, S, Bharadwaj, A, Houser, A. | | 登録日 | 2020-05-02 | | 公開日 | 2020-08-12 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Molecular principles of assembly, activation, and inhibition in epithelial sodium channel.
Elife, 9, 2020
|
|
6BKP
 
 | |
8QQ5
 
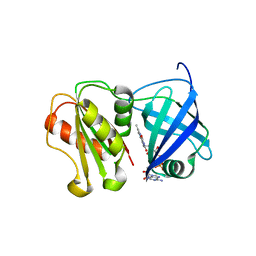 | | Structure of WT SpNox DH domain: a bacterial NADPH oxidase. | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase | | 著者 | Thepaut, M, Petit-Hartlein, I, Vermot, A, Humm, A.S, Dupeux, F, Marquez, J.A, Smith, S, Fieschi, F. | | 登録日 | 2023-10-03 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
3OOM
 
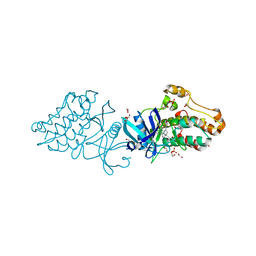 | | Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00507 | | 分子名称: | 1,2-ETHANEDIOL, 1-{3-[6-(tetrahydro-2H-pyran-4-ylamino)imidazo[1,2-b]pyridazin-3-yl]phenyl}ethanone, Activin receptor type-1, ... | | 著者 | Chaikuad, A, Sanvitale, C, Cooper, C, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Gileadi, O, Fedorov, O, Allerston, C, Krojer, T, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | 登録日 | 2010-08-31 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00507
To be Published
|
|
8TK9
 
 | | ZIG-4-INS-6 complex, tetragonal form | | 分子名称: | GLYCEROL, Probable insulin-like peptide beta-type 5, SULFATE ION, ... | | 著者 | Cheng, S, Baltrusaitis, E, Aziz, Z, Nawrocka, W.I, Ozkan, E. | | 登録日 | 2023-07-25 | | 公開日 | 2024-08-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Nematode Extracellular Protein Interactome Expands Connections between Signaling Pathways.
Biorxiv, 2024
|
|
8TKT
 
 | | ZIG-4-INS-6 complex, C-centered monoclinic form | | 分子名称: | Probable insulin-like peptide beta-type 5, Zwei Ig domain protein zig-4 | | 著者 | Cheng, S, Baltrusaitis, E, Aziz, Z, Nawrocka, W.I, Ozkan, E. | | 登録日 | 2023-07-25 | | 公開日 | 2024-08-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Nematode Extracellular Protein Interactome Expands Connections between Signaling Pathways.
Biorxiv, 2024
|
|
1Y03
 
 | | Solution structure of a recombinant type I sculpin antifreeze protein | | 分子名称: | Antifreeze peptide SS-3 | | 著者 | Kwan, A.H.Y, Fairley, K, Anderberg, P.I, Liew, C.W, Harding, M.M, Mackay, J.P. | | 登録日 | 2004-11-14 | | 公開日 | 2005-03-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a recombinant type I sculpin antifreeze protein
Biochemistry, 44, 2005
|
|
7Q16
 
 | | Human 14-3-3 zeta fused to the BAD peptide including phosphoserine-74 | | 分子名称: | 14-3-3 protein zeta/delta,Bcl2-associated agonist of cell death | | 著者 | Sluchanko, N.N, Tugaeva, K.V, Gushchin, I, Remeeva, A, Kovalev, K, Cooley, R.B. | | 登録日 | 2021-10-18 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.356 Å) | | 主引用文献 | Crystal structure of human 14-3-3 zeta complexed with the noncanonical phosphopeptide from proapoptotic BAD.
Biochem.Biophys.Res.Commun., 583, 2021
|
|
5N1T
 
 | | Crystal structure of complex between flavocytochrome c and copper chaperone CopC from T. paradoxus | | 分子名称: | COPPER (II) ION, CopC, Cytochrome C, ... | | 著者 | Osipov, E.M, Lilina, A.V, Tikhonova, T.V, Tsallagov, S.I, Popov, V.O. | | 登録日 | 2017-02-06 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of the flavocytochrome c sulfide dehydrogenase associated with the copper-binding protein CopC from the haloalkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio paradoxusARh 1.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5AGQ
 
 | | Solution structure of the TAM domain of human TIP5 BAZ2A involved in epigenetic regulation of rRNA genes | | 分子名称: | BROMODOMAIN ADJACENT TO ZINC FINGER DOMAIN PROTEIN 2A | | 著者 | Anosova, I, Melnik, S, Tripsianes, K, Kateb, F, Grummt, I, Sattler, M. | | 登録日 | 2015-02-03 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Novel RNA Binding Surface of the Tam Domain of Tip5/Baz2A Mediates Epigenetic Regulation of Rrna Genes.
Nucleic Acids Res., 43, 2015
|
|
8CM7
 
 | | W-formate dehydrogenase M405A from Desulfovibrio vulgaris | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CHLORIDE ION, ... | | 著者 | Vilela-Alves, G, Mota, C, Oliveira, A.R, Manuel, R.R, Pereira, I.C, Romao, M.J. | | 登録日 | 2023-02-17 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (2.117 Å) | | 主引用文献 | An allosteric redox switch involved in oxygen protection in a CO 2 reductase.
Nat.Chem.Biol., 20, 2024
|
|
8QWP
 
 | | Structure of p53 cancer mutant Y236C | | 分子名称: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, L(+)-TARTARIC ACID, ... | | 著者 | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | 登録日 | 2023-10-19 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
2JHQ
 
 | | Crystal structure of Uracil DNA-glycosylase from Vibrio cholerae | | 分子名称: | CHLORIDE ION, URACIL DNA-GLYCOSYLASE | | 著者 | Raeder, I.L.U, Moe, E, Willassen, N.P, Smalas, A.O, Leiros, I. | | 登録日 | 2007-02-23 | | 公開日 | 2008-06-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of Uracil-DNA N-Glycosylase (Ung) from Vibrio Cholerae. Mapping Temperature Adaptation Through Structural and Mutational Analysis.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7UQW
 
 | | PCC6803 Cyanophycinase S132DAP covalently bound to cyanophycin dimer | | 分子名称: | (2~{S})-4-[[(2~{S})-5-[[azanyl($l^{4}-azanylidene)methyl]amino]-1-$l^{1}-oxidanyl-1-oxidanylidene-pentan-2-yl]amino]-2-$l^{2}-azanyl-4-oxidanylidene-butanoic acid, Cyanophycinase, FORMAMIDE, ... | | 著者 | Sharon, I, Schmeing, T.M. | | 登録日 | 2022-04-20 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The structure of cyanophycinase in complex with a cyanophycin degradation intermediate.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
8QHS
 
 | | Cryo-EM structure of the monocin tail-tube, MttP. | | 分子名称: | Antigen A | | 著者 | Nadejda, S, Lichtenstein, R, Schlussel, S, Azulay, G, Borovok, I, Holdengraber, V, Elad, N, Wolf, S.G, Zalk, R, Zarivach, R, Frank, G.A, Herskovits, A.A. | | 登録日 | 2023-09-10 | | 公開日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Tailocin cell factories
To Be Published
|
|
1M7L
 
 | | Solution Structure of the Coiled-Coil Trimerization Domain from Lung Surfactant Protein D | | 分子名称: | Pulmonary surfactant-associated protein D | | 著者 | Kovacs, H, O'Donoghue, S.I, Hoppe, H.-J, Comfort, D, Reid, K.B.M, Campbell, I.D, Nilges, M. | | 登録日 | 2002-07-22 | | 公開日 | 2002-11-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the coiled-coil trimerization domain from lung surfactant protein D
J.BIOMOL.NMR, 24, 2002
|
|
6OOA
 
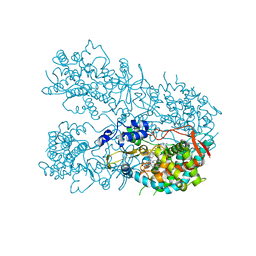 | | Human CYP3A4 bound to a drug substrate | | 分子名称: | (3aS,4R,5S,6R,8R,9R,9aR,10R)-6-ethyl-5-hydroxy-4,6,9,10-tetramethyl-1-oxodecahydro-3a,9-propanocyclopenta[8]annulen-8-yl [(5-amino-1H-1,2,4-triazol-3-yl)sulfanyl]acetate, Cytochrome P450 3A4, GLYCEROL, ... | | 著者 | Sevrioukova, I.F. | | 登録日 | 2019-04-22 | | 公開日 | 2019-09-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Structural Insights into the Interaction of Cytochrome P450 3A4 with Suicide Substrates: Mibefradil, Azamulin and 6',7'-Dihydroxybergamottin.
Int J Mol Sci, 20, 2019
|
|
7NE7
 
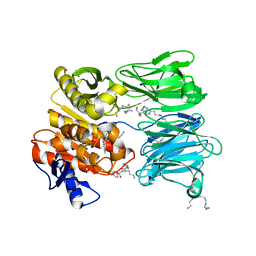 | | oligopeptidase B from S. proteomaculans with modified hinge region in complex with N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide | | 分子名称: | N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide, Oligopeptidase B, SPERMINE | | 著者 | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | 登録日 | 2021-02-03 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The Crystal Structure of N alpha-p-tosyl-lysyl Chloromethylketone-Bound Oligopeptidase B from Serratia Proteamaculans Revealed a New Type of Inhibitor Binding
Crystals, 11, 2021
|
|
7UPS
 
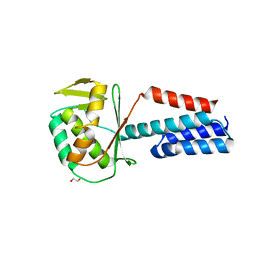 | |
1MA9
 
 | | Crystal structure of the complex of human vitamin D binding protein and rabbit muscle actin | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Alpha Skeletal Muscle, ... | | 著者 | Verboven, C, Bogaerts, I, Waelkens, E, Rabijns, A, Van Baelen, H, Bouillon, R, De Ranter, C. | | 登録日 | 2002-08-02 | | 公開日 | 2003-02-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Actin-DBP: the perfect structural fit?
Acta Crystallogr.,Sect.D, 59, 2003
|
|
7R5Y
 
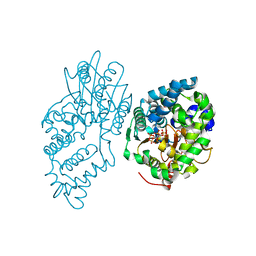 | |
6CMZ
 
 | | 2.3 Angstrom Resolution Crystal Structure of Dihydrolipoamide Dehydrogenase from Burkholderia cenocepacia in Complex with FAD and NAD | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, D-MALATE, ... | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-03-06 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | 2.3 Angstrom Resolution Crystal Structure of Dihydrolipoamide Dehydrogenase from Burkholderia cenocepacia in Complex with FAD and NAD.
To Be Published
|
|
6FPJ
 
 | | Structure of the AMPAR GluA3 N-terminal domain bound to phosphate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Herguedas, B, Garcia-Nafria, J, Greger, I. | | 登録日 | 2018-02-09 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Druggability Simulations and X-Ray Crystallography Reveal a Ligand-Binding Site in the GluA3 AMPA Receptor N-Terminal Domain.
Structure, 27, 2019
|
|
7PS7
 
 | |
7PS5
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-47 Fab | | 分子名称: | Beta-47 Fab heavy chain, Beta-47 Fab light chain, Spike protein S1, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
