2Y4P
 
 | |
2XQA
 
 | | Pentameric ligand gated ion channel GLIC in complex with tetrabutylantimony (TBSb) | | Descriptor: | ANTIMONY (III) ION, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
4QPG
 
 | | Crystal structure of empty hepatitis A virus | | Descriptor: | CHLORIDE ION, Capsid protein VP0, Capsid protein VP1, ... | | Authors: | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
4QX0
 
 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QST
 
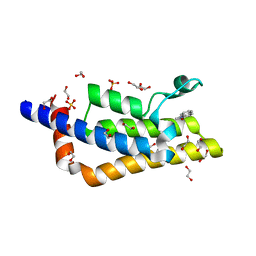 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with 1-methylquinolin 2-one | | Descriptor: | 1,2-ETHANEDIOL, 1-METHYLQUINOLIN-2(1H)-ONE, ATPase family AAA domain-containing protein 2, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
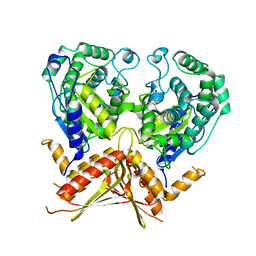
3AGD
 
 | |
2XGS
 
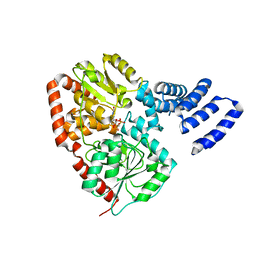 | | XcOGT in complex with C-UDP | | Descriptor: | 5'-O-[(S)-HYDROXY(PHOSPHONOMETHYL)PHOSPHORYL]URIDINE, XCOGT | | Authors: | Dorfmueller, H.C, Borodkin, V.S, Blair, D.E, Pathak, S, Navratilova, I, van Aalten, D.M. | | Deposit date: | 2010-06-07 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Substrate and Product Analogues as Human O-Glcnac Transferase Inhibitors.
Amino Acids, 40, 2011
|
|
3CZD
 
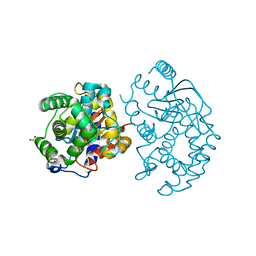 | | Crystal structure of human glutaminase in complex with L-glutamate | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutaminase kidney isoform, ... | | Authors: | Karlberg, T, Welin, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wikstrom, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4R7K
 
 | | 1.88 Angstrom Resolution Crystal Structure of Hypothetical Protein jhp0584 from Helicobacter pylori. | | Descriptor: | Hypothetical protein jhp0584 | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Hypothetical Protein jhp0584 from Helicobacter pylori.
To be Published
|
|
2XKR
 
 | | Crystal Structure of Mycobacterium tuberculosis CYP142: A novel cholesterol oxidase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 142, TETRAETHYLENE GLYCOL | | Authors: | Driscoll, M, McLean, K.J, Levy, C.W, Lafite, P, Mast, N, Pikuleva, I.A, Rigby, S.E.J, Leys, D, Munro, A.W. | | Deposit date: | 2010-07-12 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and Biochemical Characterization of Mycobacterium Tuberculosis Cyp142: Evidence for Multiple Cholesterol 27-Hydroxylase Activities in a Human Pathogen.
J.Biol.Chem., 285, 2010
|
|
3CZG
 
 | | Crystal Structure Analysis of Sucrose hydrolase (SUH)-glucose complex | | Descriptor: | Sucrose hydrolase, alpha-D-glucopyranose | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and mutagenesis of sucrose hydrolase from Xanthomonas axonopodis pv. glycines: insight into the exclusively hydrolytic amylosucrase fold.
J.Mol.Biol., 380, 2008
|
|
2XCB
 
 | | Crystal structure of PcrH in complex with the chaperone binding region of PopD | | Descriptor: | NITRATE ION, PEPD, REGULATORY PROTEIN PCRH | | Authors: | Job, V, Mattei, P.-J, Lemaire, D, Attree, I, Dessen, A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Chaperone Recognition of Type III Secretion System Minor Translocator Proteins.
J.Biol.Chem., 285, 2010
|
|
4QSP
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with acetyl-lysine | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, N(6)-ACETYLLYSINE, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
4QSV
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with thymidine | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, SULFATE ION, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
4S3K
 
 | |
2XWX
 
 | | Vibrio cholerae colonization factor GbpA crystal structure | | Descriptor: | GLCNAC-BINDING PROTEIN A | | Authors: | Wong, E, Vaaje-Kolstad, G, Ghosh, A, Guerrero, R.H, Konarev, P.V, Ibrahim, A.F.M, Svergun, D.I, Eijsink, V.G.H, Chatterjee, N.S, van Aalten, D.M.F. | | Deposit date: | 2010-11-06 | | Release date: | 2011-11-16 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Vibrio Cholerae Colonization Factor Gbpa Possesses a Modular Structure that Governs Binding to Different Host Surfaces.
Plos Pathog., 8, 2012
|
|
4RLN
 
 | | Hen egg-white lysozyme solved from serial crystallography at a synchrotron source, data processed with nXDS | | Descriptor: | Lysozyme C | | Authors: | Botha, S, Nass, K, Barends, T, Kabsch, W, Latz, B, Dworkowski, F, Foucar, L, Panepucci, E, Wang, M, Shoeman, R, Schlichting, I, Doak, R.B. | | Deposit date: | 2014-10-17 | | Release date: | 2015-02-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Room-temperature serial crystallography at synchrotron X-ray sources using slowly flowing free-standing high-viscosity microstreams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2Y2C
 
 | | crystal structure of AmpD Apoenzyme | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
3CQV
 
 | | Crystal structure of Reverb beta in complex with heme | | Descriptor: | Nuclear receptor subfamily 1 group D member 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, X, Dong, A, Pardee, K.I, Reinking, J, Krause, H, Schuetz, A, Zhang, R, Cui, H, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Savchenko, A, Botchkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-03 | | Release date: | 2008-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBbeta.
Plos Biol., 7, 2009
|
|
4RMI
 
 | | Human Sirt2 in complex with SirReal1 and Ac-Lys-OTC peptide | | Descriptor: | Ac-Lys-OTC peptide, N-(5-benzyl-1,3-thiazol-2-yl)-2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetamide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4RNI
 
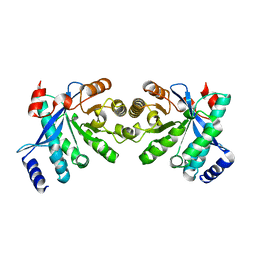 | | PaMorA dimeric phosphodiesterase. apo form | | Descriptor: | Motility regulator | | Authors: | Phippen, C.W, Tews, I. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Formation and dimerization of the phosphodiesterase active site of the Pseudomonas aeruginosa MorA, a bi-functional c-di-GMP regulator.
Febs Lett., 588, 2014
|
|
2XF1
 
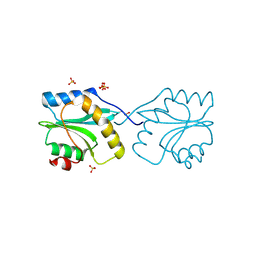 | | Crystal structure of Plasmodium falciparum actin depolymerization factor 1 | | Descriptor: | COFILIN ACTIN-DEPOLYMERIZING FACTOR HOMOLOG 1, SULFATE ION | | Authors: | Singh, B.K, Sattler, J.M, Huttu, J, Chatterjee, M, Schueler, H, Kursula, I. | | Deposit date: | 2010-05-20 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures Explain Functional Differences in the Two Actin Depolymerization Factors of the Malaria Parasite.
J.Biol.Chem., 286, 2011
|
|
3B30
 
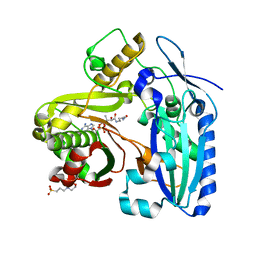 | | Crystal Structure of F. graminearum TRI101 complexed with Ethyl Coenzyme A | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Ethyl Coenzyme A, Trichothecene 3-O-acetyltransferase | | Authors: | Garvey, G.S, Rayment, I. | | Deposit date: | 2007-10-19 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Functional Characterization of the TRI101 Trichothecene 3-O-Acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum: KINETIC INSIGHTS TO COMBATING FUSARIUM HEAD BLIGHT
J.Biol.Chem., 283, 2008
|
|
2V30
 
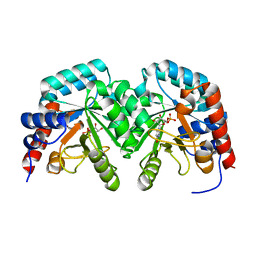 | | Human orotidine 5'-phosphate decarboxylase domain of uridine monophospate synthetase (UMPS) in complex with its product UMP. | | Descriptor: | OROTIDINE 5'-PHOSPHATE DECARBOXYLASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Moche, M, Ogg, D, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, van den Berg, S, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Orotidine 5'-Decarboxylase Domain of Human Uridine Monophosphate Synthetase (Umps)
To be Published
|
|
3B2S
 
 | | Crystal Structure of F. graminearum TRI101 complexed with Coenzyme A and Deoxynivalenol | | Descriptor: | (3alpha,7alpha)-3,7,15-trihydroxy-12,13-epoxytrichothec-9-en-8-one, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, COENZYME A, ... | | Authors: | Garvey, G.S, Rayment, I. | | Deposit date: | 2007-10-19 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and Functional Characterization of the TRI101 Trichothecene 3-O-Acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum: KINETIC INSIGHTS TO COMBATING FUSARIUM HEAD BLIGHT
J.Biol.Chem., 283, 2008
|
|
