7RDF
 
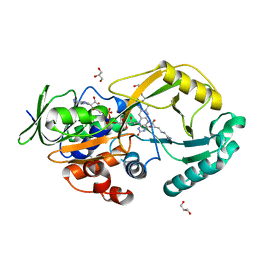 | | Crystal structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F co-crystallized in the presence of D-arginine | | Descriptor: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, FAD-dependent catabolic D-arginine dehydrogenase DauA, ... | | Authors: | Reis, R.A.G, Iyer, A, Agniswamy, A, Weber, I.T, Gadda, G. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Discovery of a new flavin N5-adduct in a tyrosine to phenylalanine variant of d-Arginine dehydrogenase.
Arch.Biochem.Biophys., 715, 2021
|
|
4A8F
 
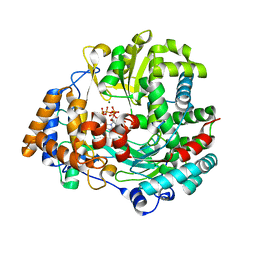 | | Non-Catalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial dsRNA virus phi6 from De Novo Initiation to Elongation | | Descriptor: | 5'-D(*DAP*GP*CP*GP)-3', ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wright, S, Poranen, M.M, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Noncatalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial Double-Stranded RNA Virus Phi6 from De Novo Initiation to Elongation.
J.Virol., 86, 2012
|
|
4AH9
 
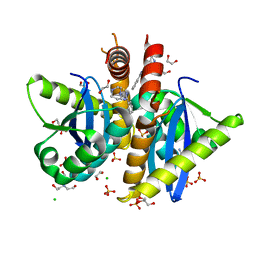 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-PHENYL-1,2,4-THIADIAZOL-5-YL)-1,4-DIAZEPANE, CHLORIDE ION, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-06 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
2QXW
 
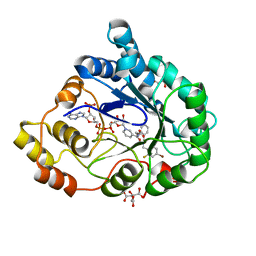 | | Perdeuterated alr2 in complex with idd594 | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Blakeley, M.P, Ruiz, F, Cachau, R, Hazemann, I, Meilleur, F, Mitschler, A, Ginell, S, Afonine, P, Ventura, O, Cousido-Siah, A, Joachimiak, A, Myles, D, Podjarny, A. | | Deposit date: | 2007-08-13 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Quantum model of catalysis based on a mobile proton revealed by subatomic x-ray and neutron diffraction studies of h-aldose reductase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5JXP
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 in alternate conformation | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CALCIUM ION, CHLORIDE ION | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
1HRQ
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE REDUCED HIGH-POTENTIAL IRON-SULFUR PROTEIN FROM CHROMATIUM VINOSUM THROUGH NMR | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Banci, L, Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | Deposit date: | 1995-01-17 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the reduced high-potential iron-sulfur protein from Chromatium vinosum through NMR.
Biochemistry, 34, 1995
|
|
1HRR
 
 | | THE THREE DIMENSIONAL STRUCTURE OF THE REDUCED HIGH POTENTIAL IRON-SULFUR PROTEIN FROM CHROMATIUM VINOSUM THROUGH NMR | | Descriptor: | IRON/SULFUR CLUSTER, REDUCED HIGH POTENTIAL IRON SULFUR PROTEIN | | Authors: | Banci, L, Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | Deposit date: | 1995-01-17 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the reduced high-potential iron-sulfur protein from Chromatium vinosum through NMR.
Biochemistry, 34, 1995
|
|
7WWO
 
 | |
1HK1
 
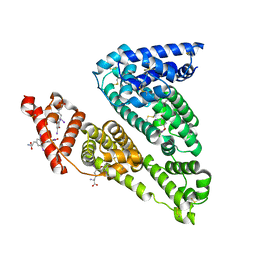 | | HUMAN SERUM ALBUMIN COMPLEXED WITH THYROXINE (3,3',5,5'-TETRAIODO-L-THYRONINE) | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, SERUM ALBUMIN | | Authors: | Petitpas, I, Petersen, C.E, Ha, C.E, Bhattacharya, A.A, Zunszain, P.A, Ghuman, J, Bhagavan, N.V, Curry, S. | | Deposit date: | 2003-03-05 | | Release date: | 2003-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis of Albumin-Thyroxine Interactions and Familial Dysalbuminemic Hyperthyroxinemia
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1HK3
 
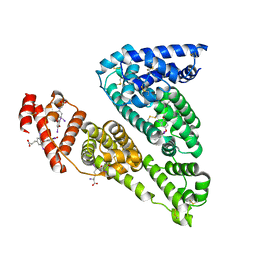 | | Human serum albumin mutant r218p complexed with thyroxine (3,3',5,5'-tetraiodo-l-thyronine) | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, SERUM ALBUMIN | | Authors: | Petitpas, I, Petersen, C.E, Ha, C.E, Bhattacharya, A.A, Zunszain, P.A, Ghuman, J, Bhagavan, N.V, Curry, S. | | Deposit date: | 2003-03-05 | | Release date: | 2003-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Albumin-Thyroxine Interactions and Familial Dysalbuminemic Hyperthyroxinemia
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1HK2
 
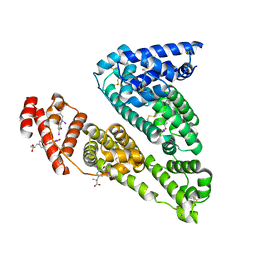 | | HUMAN SERUM ALBUMIN MUTANT R218H COMPLEXED WITH THYROXINE (3,3',5,5'-TETRAIODO-L-THYRONINE) | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, SERUM ALBUMIN | | Authors: | Petitpas, I, Petersen, C.E, Ha, C.E, Bhattacharya, A.A, Zunszain, P.A, Ghuman, J, Bhagavan, N.V, Curry, S. | | Deposit date: | 2003-03-05 | | Release date: | 2003-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Albumin-Thyroxine Interactions and Familial Dysalbuminemic Hyperthyroxinemia
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5KHI
 
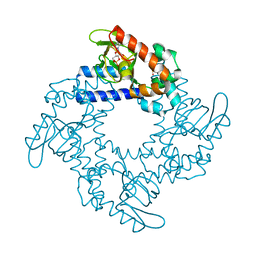 | | HCN2 CNBD in complex with purine riboside-3', 5'-cyclic monophosphate (cPuMP) | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2, Purine riboside-3',5'-cyclic monophosphate | | Authors: | Ng, L.C.T, Putrenko, I, Baronas, V, Van Petegem, F, Accili, E.A. | | Deposit date: | 2016-06-14 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cyclic Purine and Pyrimidine Nucleotides Bind to the HCN2 Ion Channel and Variably Promote C-Terminal Domain Interactions and Opening.
Structure, 24, 2016
|
|
7RLT
 
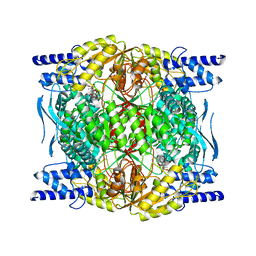 | | Structure of ligand-free ALDH1L1 (10-formyltetrahydrofolate dehydrogenase) | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Cytosolic 10-formyltetrahydrofolate dehydrogenase | | Authors: | Tsybovsky, Y, Sereda, V, Golczak, M, Krupenko, N.I, Krupenko, S.A. | | Deposit date: | 2021-07-26 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of putative tumor suppressor ALDH1L1.
Commun Biol, 5, 2022
|
|
7X0C
 
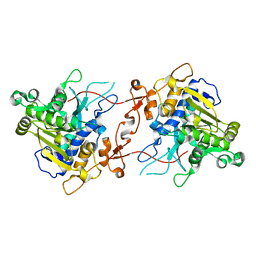 | | Crystal structure of phospholipase A1, AtDSEL | | Descriptor: | Phospholipase A1-IIgamma | | Authors: | Heo, Y, Lee, I, Moon, S, Lee, W. | | Deposit date: | 2022-02-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79909515 Å) | | Cite: | Crystal Structures of the Plant Phospholipase A1 Proteins Reveal a Unique Dimerization Domain.
Molecules, 27, 2022
|
|
7RM1
 
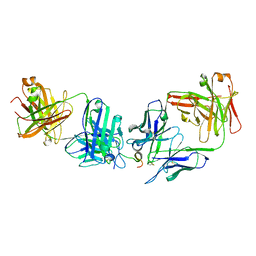 | |
5CM3
 
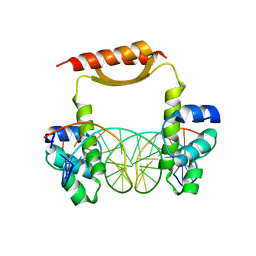 | | Crystal Structure of KorA, a plasmid-encoded, global transcription regulator | | Descriptor: | 5'-D(CP*CP*AP*AP*GP*TP*TP*TP*AP*GP*CP*TP*AP*AP*AP*CP*TP*TP*GP*GP*)-3', TrfB transcriptional repressor protein | | Authors: | White, S.A, Hyde, E.I, Rajasekar, K.V. | | Deposit date: | 2015-07-16 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
7RLX
 
 | |
4AHS
 
 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1-BENZOFURAN-7-CARBOXYLIC ACID, ACETATE ION, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
5KDN
 
 | | ZmpB metallopeptidase from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, F5/8 type C domain protein, ZINC ION | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3FX8
 
 | | Distinct recognition of three-way DNA junctions by a thioester variant of a metallo-supramolecular cylinder ('helicate') | | Descriptor: | (5'-D(*CP*GP*TP*AP*CP*G)-3', 4,4'-sulfanediylbis{N-[(1E)-pyridin-2-ylmethylidene]aniline}, FE (II) ION | | Authors: | Boer, D.R, Uson, I, Coll, M. | | Deposit date: | 2009-01-20 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Self-Assembly of Functionalizable Two-Component 3D DNA Arrays through the Induced Formation of DNA Three-Way-Junction Branch Points by Supramolecular Cylinders.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
5KG9
 
 | |
5KHK
 
 | | HCN2 CNBD in complex with 2-aminopurine riboside-3', 5'-cyclic monophosphate (2-NH2-cPuMP) | | Descriptor: | 2-Aminopurine riboside-3',5'-cyclic monophosphate, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Ng, L.C.T, Putrenko, I, Baronas, V, Van Petegem, F, Accili, E.A. | | Deposit date: | 2016-06-14 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Cyclic Purine and Pyrimidine Nucleotides Bind to the HCN2 Ion Channel and Variably Promote C-Terminal Domain Interactions and Opening.
Structure, 24, 2016
|
|
5KHJ
 
 | | HCN2 CNBD in complex with uridine-3', 5'-cyclic monophosphate (cUMP) | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2, Uridine-3',5'-cyclic monophosphate | | Authors: | Ng, L.C.T, Putrenko, I, Baronas, V, Van Petegem, F, Accili, E.A. | | Deposit date: | 2016-06-14 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Cyclic Purine and Pyrimidine Nucleotides Bind to the HCN2 Ion Channel and Variably Promote C-Terminal Domain Interactions and Opening.
Structure, 24, 2016
|
|
1H5D
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (0-11% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, PEROXIDASE C1A, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-21 | | Release date: | 2002-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
4A66
 
 | | Mutations in the neighbourhood of CotA-laccase trinuclear site: D116A mutant | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, PEROXIDE ION, ... | | Authors: | Silva, C.S, Lindley, P.F, Bento, I. | | Deposit date: | 2011-10-31 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Role of Asp116 in the Reductive Cleavage of Dioxygen to Water in Cota Laccase: Assistance During the Proton Transfer Mechanism
Acta Crystallogr.,Sect.D, 68, 2012
|
|
