4GEX
 
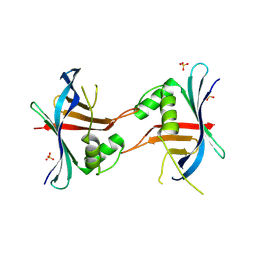 | |
6QWX
 
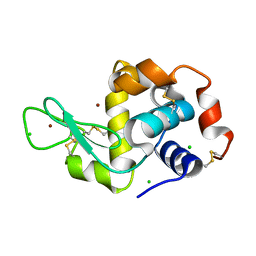 | | HEWL lysozyme, crystallized from NiCl2 solution | | Descriptor: | CHLORIDE ION, Lysozyme C, NICKEL (II) ION | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
3LXA
 
 | | Interconversion of Human Lysosomal Enzyme Specificities | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, ... | | Authors: | Tomasic, I.B, Metcalf, M.C, Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2010-02-25 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Interconversion of the specificities of human lysosomal enzymes associated with Fabry and Schindler diseases.
J.Biol.Chem., 285, 2010
|
|
7UF9
 
 | | CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-{[(2R)-1-oxo-3-phenyl-1-{[2-(pyridin-4-yl)ethyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Interaction of CYP3A4 with Rationally Designed Ritonavir Analogues: Impact of Steric Constraints Imposed on the Heme-Ligating Group and the End-Pyridine Attachment.
Int J Mol Sci, 23, 2022
|
|
2FX8
 
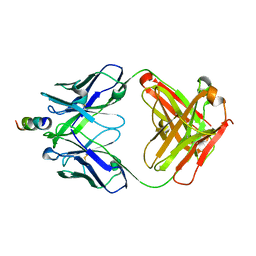 | | Crystal structure of hiv-1 neutralizing human fab 4e10 in complex with an aib-induced peptide encompassing the 4e10 epitope on gp41 | | Descriptor: | Fab 4E10, Fragment of HIV glycoprotein (GP41) | | Authors: | Cardoso, R.M.F, Brunel, F.M, Ferguson, S, Burton, D.R, Dawson, P.E, Wilson, I.A. | | Deposit date: | 2006-02-03 | | Release date: | 2006-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10.
J.Mol.Biol., 365, 2007
|
|
7UFE
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, GLYCEROL, N-[(2S)-1-{[(2R)-1-oxo-3-phenyl-1-{[3-(pyridin-3-yl)propyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]pyridine-3-carboxamide, ... | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interaction of CYP3A4 with Rationally Designed Ritonavir Analogues: Impact of Steric Constraints Imposed on the Heme-Ligating Group and the End-Pyridine Attachment.
Int J Mol Sci, 23, 2022
|
|
6QSZ
 
 | | Crystal structure of the Sir4 H-BRCT domain in complex with Esc1 pS1450 peptide | | Descriptor: | CHLORIDE ION, Regulatory protein SIR4, Silent chromatin protein ESC1 | | Authors: | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | Deposit date: | 2019-02-22 | | Release date: | 2019-09-18 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
8SI6
 
 | | Cryo-EM structure of TRPM7 in MSP2N2 nanodisc in complex with agonist naltriben in closed state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (4bS,8R,8aS,14bR)-7-(cyclopropylmethyl)-5,6,7,8,9,14b-hexahydro-8aH-4,8-methanobis[1]benzofuro[3,2-e:2',3'-g]isoquinoline-1,8a-diol, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
7UFA
 
 | | CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-{[(2R)-1-oxo-3-phenyl-1-{[3-(pyridin-4-yl)propyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interaction of CYP3A4 with Rationally Designed Ritonavir Analogues: Impact of Steric Constraints Imposed on the Heme-Ligating Group and the End-Pyridine Attachment.
Int J Mol Sci, 23, 2022
|
|
8SI5
 
 | | Cryo-EM structure of TRPM7 in MSP2N2 nanodisc in complex with agonist naltriben in open state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (4bS,8R,8aS,14bR)-7-(cyclopropylmethyl)-5,6,7,8,9,14b-hexahydro-8aH-4,8-methanobis[1]benzofuro[3,2-e:2',3'-g]isoquinoline-1,8a-diol, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8D4R
 
 | |
7UFB
 
 | | CYP3A4 bound to an inhibitor | | Descriptor: | (2R)-3-phenyl-2-({(2S)-3-phenyl-2-[2-(pyridin-3-yl)acetamido]propyl}sulfanyl)-N-[(pyridin-3-yl)methyl]propanamide, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Interaction of CYP3A4 with Rationally Designed Ritonavir Analogues: Impact of Steric Constraints Imposed on the Heme-Ligating Group and the End-Pyridine Attachment.
Int J Mol Sci, 23, 2022
|
|
8D50
 
 | |
8SI4
 
 | | Cryo-EM structure of TRPM7 N1098Q mutant in GDN detergent in open state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, CALCIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
3LXB
 
 | | Interconversion of Human Lysosomal Enzyme Specificities | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, ... | | Authors: | Tomasic, I.B, Metcalf, M.C, Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2010-02-25 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Interconversion of the specificities of human lysosomal enzymes associated with Fabry and Schindler diseases.
J.Biol.Chem., 285, 2010
|
|
3LXC
 
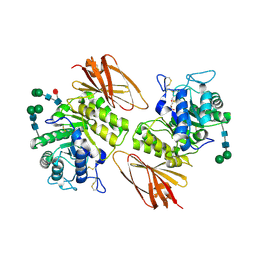 | | Interconversion of Human Lysosomal Enzyme Specificities | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, GLYCEROL, ... | | Authors: | Tomasic, I.B, Metcalf, M.C, Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2010-02-25 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Interconversion of the specificities of human lysosomal enzymes associated with Fabry and Schindler diseases.
J.Biol.Chem., 285, 2010
|
|
8SI7
 
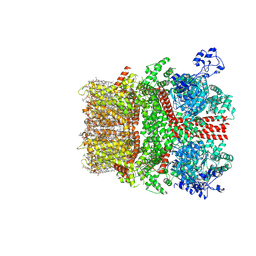 | | Cryo-EM structure of TRPM7 in GDN detergent in complex with inhibitor VER155008 in closed state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8SIA
 
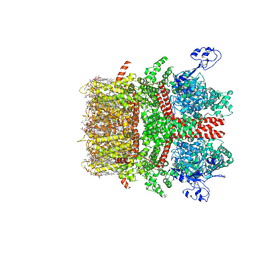 | | Cryo-EM structure of TRPM7 N1098Q mutant in GDN detergent in complex with inhibitor NS8593 in closed state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, CALCIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8SI2
 
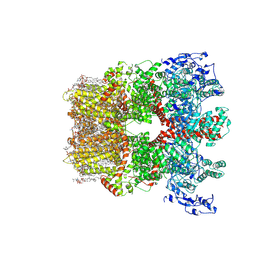 | | Cryo-EM structure of TRPM7 in MSP2N2 nanodisc in apo state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, CHOLESTEROL, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
1RDJ
 
 | | MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH BETA-METHYL-L-FUCOPYRANOSIDE | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-C, ... | | Authors: | Ng, K.K.-S, Drickamer, K, Weis, W.I. | | Deposit date: | 1995-09-05 | | Release date: | 1996-03-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of monosaccharide recognition by rat liver mannose-binding protein.
J.Biol.Chem., 271, 1996
|
|
8D53
 
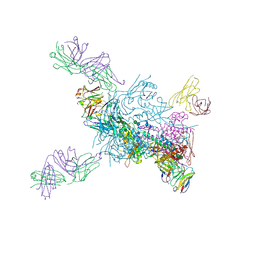 | |
8DHR
 
 | | An ester mutant of SfGFP | | Descriptor: | Green fluorescent protein | | Authors: | Reddi, R, Valiyaveetil, F.I. | | Deposit date: | 2022-06-28 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A facile approach for incorporating tyrosine esters to probe ion-binding sites and backbone hydrogen bonds.
J.Biol.Chem., 300, 2023
|
|
8SI3
 
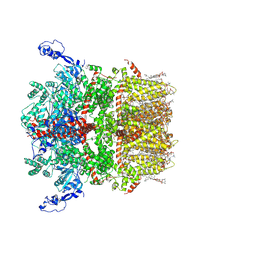 | | Cryo-EM structure of TRPM7 in GDN detergent in apo state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, CALCIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8SI8
 
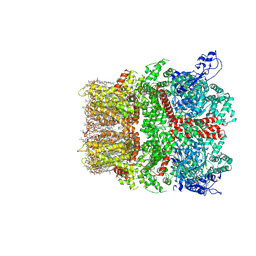 | | Cryo-EM structure of TRPM7 N1098Q mutant in GDN detergent in complex with inhibitor VER155008 in closed state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
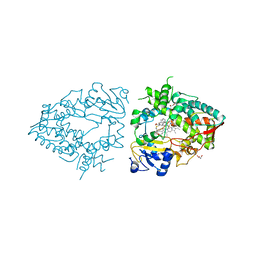
7UQW
 
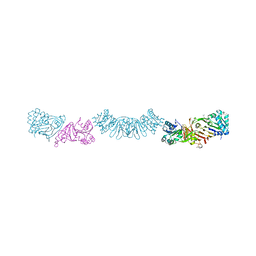 | | PCC6803 Cyanophycinase S132DAP covalently bound to cyanophycin dimer | | Descriptor: | (2~{S})-4-[[(2~{S})-5-[[azanyl($l^{4}-azanylidene)methyl]amino]-1-$l^{1}-oxidanyl-1-oxidanylidene-pentan-2-yl]amino]-2-$l^{2}-azanyl-4-oxidanylidene-butanoic acid, Cyanophycinase, FORMAMIDE, ... | | Authors: | Sharon, I, Schmeing, T.M. | | Deposit date: | 2022-04-20 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of cyanophycinase in complex with a cyanophycin degradation intermediate.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
