7QVR
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-37 (G206S, R207T, D210S) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | Authors: | Loch, J.I, Kadziolka, K, Jaskolski, M. | | Deposit date: | 2022-01-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QY6
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, wild type (WT EcAIII) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | Authors: | Loch, J.I, Klonecka, A, Kadziolka, K, Bonarek, P, Barciszewski, J, Imiolczyk, B, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2022-01-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6HPN
 
 | | Crystal structure of Borrelia spielmanii WP_012665240 (BSA64) - an orthologous protein to B. burgdorferi BBA64 | | Descriptor: | Antigen, P35 | | Authors: | Brangulis, K, Akopjana, I, Kazaks, A, Tars, K. | | Deposit date: | 2018-09-21 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the outer surface proteins from Borrelia burgdorferi paralogous gene family 54 that are thought to be the key players in the pathogenesis of Lyme disease.
J.Struct.Biol., 210, 2020
|
|
7QQ8
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-8 (G206Y, R207Q, D210P, S211T) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, SODIUM ION | | Authors: | Loch, J.I, Kadziolka, K, Jaskolski, M. | | Deposit date: | 2022-01-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6H82
 
 | | Cryo-EM structure of the archaeal extremophilic internal membrane containing Haloarcula hispanica icosahedral virus 2 (HHIV-2) at 3.78 Angstroms resolution. | | Descriptor: | GPS III, Uncharacterized protein, VP16 (vertex complex), ... | | Authors: | Abrescia, N.G, Santos-Perez, I, Charro, D. | | Deposit date: | 2018-08-01 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural basis for assembly of vertical single beta-barrel viruses.
Nat Commun, 10, 2019
|
|
1BS5
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM | | Descriptor: | PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ZINC ION | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
5WEO
 
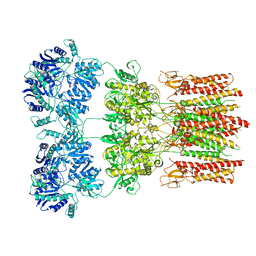 | | Activated GluA2 complex bound to glutamate, cyclothiazide, and STZ in digitonin | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit chimera | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-07-10 | | Release date: | 2017-08-02 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Channel opening and gating mechanism in AMPA-subtype glutamate receptors.
Nature, 549, 2017
|
|
1BS8
 
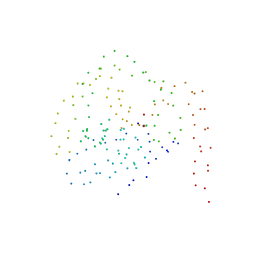 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | Descriptor: | PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
6WPM
 
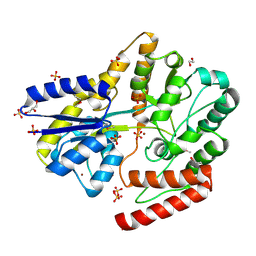 | | Crystal structure of a putative oligosaccharide periplasmic-binding protein from Synechococcus sp. MITs9220 in complex with zinc | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Ford, B.A, Michie, K.A, Paulsen, I.T, Mabbutt, B.C, Shah, B.S. | | Deposit date: | 2020-04-27 | | Release date: | 2021-05-12 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Novel functional insights into a modified sugar-binding protein from Synechococcus MITS9220.
Sci Rep, 12, 2022
|
|
5WLO
 
 | | a novel 13-ring macrocyclic HIV-1 protease inhibitors involving the P1'-P2' ligands | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[(7E)-13-methoxy-1,1-dioxo-1,4,5,6,9,11-hexahydro-10,1lambda~6~,2-benzoxathiazacyclotridecin-2(3H)-yl]-1-phenylbutan-2-yl}carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-07-27 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Design, synthesis, X-ray studies, and biological evaluation of novel macrocyclic HIV-1 protease inhibitors involving the P1'-P2' ligands.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6OVA
 
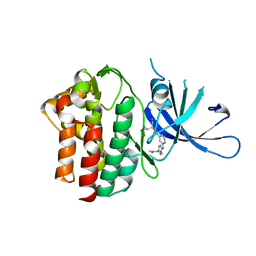 | | Crystal Structure of TYK2 with novel pyrrolidinone inhibitor | | Descriptor: | 6-({4-[(3S)-3-cyano-3-cyclopropyl-2-oxopyrrolidin-1-yl]pyridin-2-yl}amino)-N,N-dimethylpyridine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Skene, R.J, Hoffman, I.D. | | Deposit date: | 2019-05-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Efficient synthesis of tert-butyl 3-cyano-3-cyclopropyl-2-oxopyrrolidine-4-carboxylates: Highly functionalized 2-pyrrolidinone enabling access to novel macrocyclic Tyk2 inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6HNN
 
 | | Crystal structure of wild-type IdmH, a putative polyketide cyclase from Streptomyces antibioticus | | Descriptor: | Putative polyketide cyclase IdmH | | Authors: | Drulyte, I, Obajdin, J, Trinh, C, Hemsworth, G.R, Berry, A. | | Deposit date: | 2018-09-16 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the putative cyclase IdmH from the indanomycin nonribosomal peptide synthase/polyketide synthase.
Iucrj, 6, 2019
|
|
5WQF
 
 | | Structure of fungal meroterpenoid isomerase Trt14 | | Descriptor: | CALCIUM ION, Isomerase trt14, N-PROPANOL | | Authors: | Mori, T, Iwabuchi, T, Matsuda, Y, Abe, I. | | Deposit date: | 2016-11-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Molecular basis for the unusual ring reconstruction in fungal meroterpenoid biogenesis
Nat. Chem. Biol., 13, 2017
|
|
2WMW
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1-[(2S)-4-(5-BROMO-1H-PYRAZOLO[3,4-B]PYRIDIN-4-YL)MORPHOLIN-2-YL]METHANAMINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
5WPB
 
 | | Crystal structure of fragment 3-(3-(pyridin-2-ylmethoxy)quinoxalin-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-{3-[(pyridin-2-yl)methoxy]quinoxalin-2-yl}propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Tempel, W, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-04 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
2WMT
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 2-(methylsulfanyl)-5-(thiophen-2-ylmethyl)-1H-imidazol-4-ol, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
2WMV
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1-[(2S)-4-(7H-PURIN-6-YL)MORPHOLIN-2-YL]METHANAMINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
5WQG
 
 | | Structure of fungal meroterpenoid isomerase Trt14 complexed with terretonin D | | Descriptor: | CALCIUM ION, Isomerase trt14, methyl (2S,4aR,4bS,5S,6aS,10aS,10bS,12aS)-2,4b,7,7,10a,12a-hexamethyl-12-methylidene-5-oxidanyl-1,4,6,8-tetrakis(oxidanylidene) -5,6a,9,10,10b,11-hexahydro-4aH-naphtho[1,2-h]isochromene-2-carboxylate | | Authors: | Mori, T, Matsuda, Y, Abe, I. | | Deposit date: | 2016-11-26 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the unusual ring reconstruction in fungal meroterpenoid biogenesis
Nat. Chem. Biol., 13, 2017
|
|
2WKR
 
 | | Structure of a photoactivatable Rac1 containing the Lov2 C450M Mutant | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, Y.I, Frey, D, Lungu, O.I, Jaehrig, A, Schlichting, I, Kuhlman, B, Hahn, K.M. | | Deposit date: | 2009-06-16 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Genetically Encoded Photoactivatable Rac Controls the Motility of Living Cells.
Nature, 461, 2009
|
|
3H1I
 
 | | Stigmatellin and antimycin bound cytochrome bc1 complex from chicken | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-METHYL-BUTYRIC ACID 3-(3-FORMYLAMINO-2-HYDROXY-BENZOYLAMINO)-8-HEPTYL-2,6-DIMETHYL-4,9-DIOXO-[1,5]DIOXONAN-7-YL ESTER, CARDIOLIPIN, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.I, Kim, K.K, Hung, L.W, Crofts, A.R, Berry, E.A, Kim, S.H. | | Deposit date: | 2009-04-12 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
6HV0
 
 | | IRE1 kinase/RNase in complex with imidazo[1,2-b]pyridazin-8-amine compound 33 | | Descriptor: | 6-chloranyl-3-(2~{H}-indazol-5-yl)-~{N}-propan-2-yl-imidazo[1,2-b]pyridazin-8-amine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Bayliss, R, Bhatia, C, Collins, I. | | Deposit date: | 2018-10-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Binding to an Unusual Inactive Kinase Conformation by Highly Selective Inhibitors of Inositol-Requiring Enzyme 1 alpha Kinase-Endoribonuclease.
J.Med.Chem., 62, 2019
|
|
2WMS
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE CHK1, [4-amino-2-(tert-butylamino)-1,3-thiazol-5-yl](phenyl)methanone | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
6WPY
 
 | |
5WSL
 
 | | Structural studies of keratinase from Meiothermus taiwanensis WR-220 | | Descriptor: | CALCIUM ION, SULFATE ION, keratinase | | Authors: | Ho, M.C, Wu, S.H, Chen, M.Y, Tu, I.F. | | Deposit date: | 2016-12-07 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The discovery of novel heat-stable keratinases from Meiothermus taiwanensis WR-220 and other extremophiles
Sci Rep, 7, 2017
|
|
1H9Z
 
 | |
