6PXY
 
 | |
5N0I
 
 | | Crystal structure of NDM-1 in complex with beta-mercaptoethanol - new refinement | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
6QF7
 
 | | Crystal structures of the recombinant beta-Factor XIIa protease with bound Thr-Arg and Pro-Arg substrate mimetics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XII, ... | | Authors: | Pathak, M, Mannal, R, Li, C, Bubacarr, G.K, Badraldin, K.H, Belviso, B.D, Camila, R.B, Dreveny, I, Fischer, P.M, Dekker, L.V, Oliva, M.L.V, Emsley, J. | | Deposit date: | 2019-01-09 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structures of the recombinant beta-factor XIIa protease with bound Thr-Arg and Pro-Arg substrate mimetics.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8F41
 
 | | 3-methylcrotonyl-CoA carboxylase in filament, alpha-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, alpha-subunit, beta-subunit, ... | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-10 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
5M1T
 
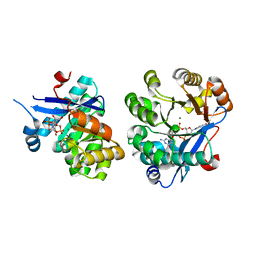 | | PaMucR Phosphodiesterase, c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, MucR Phosphodiesterase | | Authors: | Hutchin, A, Tews, I, Walsh, M.A. | | Deposit date: | 2016-10-10 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
6BPW
 
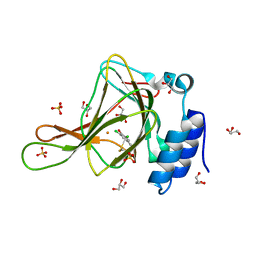 | | Crystal structure of ferrous form of the Cl2-Tyr157 human cysteine dioxygenase with both uncrosslinked and crosslinked cofactor | | Descriptor: | Cysteine dioxygenase type 1, FE (II) ION, GLYCEROL, ... | | Authors: | Liu, A, Li, J, Shin, I. | | Deposit date: | 2017-11-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Cleavage of a carbon-fluorine bond by an engineered cysteine dioxygenase.
Nat. Chem. Biol., 14, 2018
|
|
7NNK
 
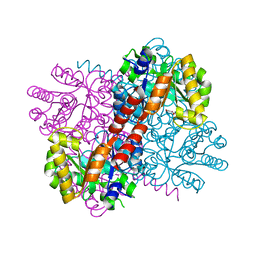 | | Crystal structure of S116A mutant of hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1 in complex with hydroxypyruvate | | Descriptor: | 3-HYDROXYPYRUVIC ACID, HpcH/HpaI aldolase, MAGNESIUM ION | | Authors: | Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-02-25 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6PS4
 
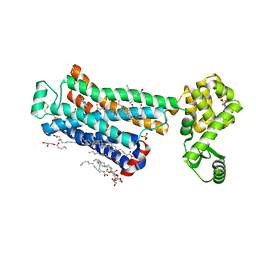 | | XFEL beta2 AR structure by ligand exchange from Timolol to ICI-118551. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3S)-1-[(7-methyl-2,3-dihydro-1H-inden-4-yl)oxy]-3-[(1-methylethyl)amino]butan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
5LWY
 
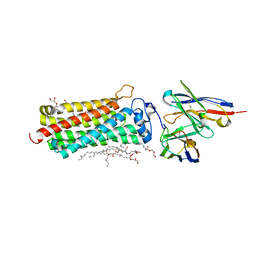 | | Revised crystal structure of the human adiponectin receptor 2 in complex with a C18 free fatty acid | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, GLYCEROL, ... | | Authors: | Leyrat, C, Vasiliauskaite-Brooks, I, Granier, S. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into adiponectin receptors suggest ceramidase activity.
Nature, 6, 2017
|
|
6PY4
 
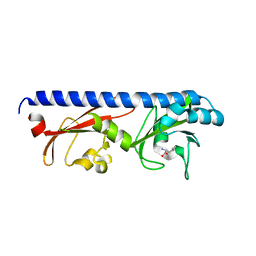 | |
1NRS
 
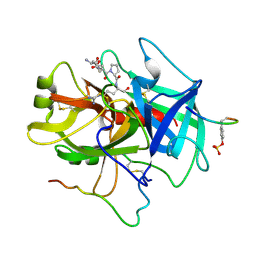 | |
8BBO
 
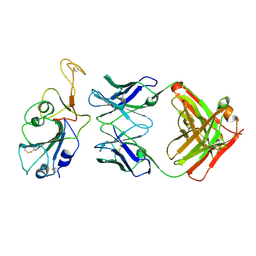 | | SARS-CoV-2 Delta-RBD complexed with BA.2-36 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IGH@ protein, Immunoglobulin kappa light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
7NNP
 
 | | Rb-loaded cryo-EM structure of the E1-ATP KdpFABC complex. | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
7NNL
 
 | | Cryo-EM structure of the KdpFABC complex in an E1-ATP conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
5NZM
 
 | | Crystal structure of UDP-glucose pyrophosphorylase from Leishmania major in complex with murrayamine-I | | Descriptor: | Murrayamine-I, UDP-glucose pyrophosphorylase | | Authors: | Cramer, J.T, Fuehring, J.I, Baruch, P, Bruetting, C, Hesse, R, Knoelker, H.-J, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2017-05-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies
Acs Catalysis, 8, 2018
|
|
5NZJ
 
 | | Crystal structure of UDP-glucose pyrophosphorylase G45Y mutant from Leishmania major in complex with UDP-glucose | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, UDP-glucose pyrophosphorylase, ... | | Authors: | Cramer, J.T, Fuehring, J.I, Baruch, P, Bruetting, C, Hesse, R, Knoelker, H.-J, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2017-05-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies
Acs Catalysis, 8, 2018
|
|
8BWO
 
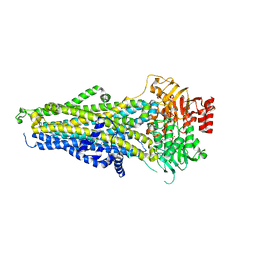 | | Cryo-EM structure of nanodisc-reconstituted human MRP4 with E1202Q mutation (outward-facing occluded conformation) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 4, MAGNESIUM ION | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BJF
 
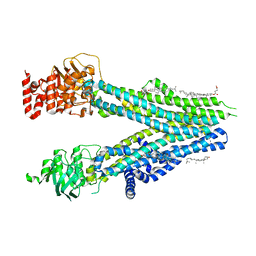 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (inward-facing conformation) | | Descriptor: | ATP-binding cassette sub-family C member 4, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
5O0S
 
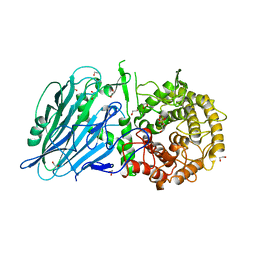 | | Crystal structure of txGH116 (beta-glucosidase from Thermoanaerobacterium xylolyticum) in complex with unreacted beta Cyclophellitol Cyclosulfate probe ME711 | | Descriptor: | (3~{a}~{S},4~{R},5~{S},6~{R},7~{R},7~{a}~{R})-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3~{a},4,5,6,7,7~{a}-hexahydrobenzo[d][1,3,2]dioxathiole-4,5,6-triol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Wu, L, Offen, W.A, Breen, I.Z, Davies, G.J. | | Deposit date: | 2017-05-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
8BWQ
 
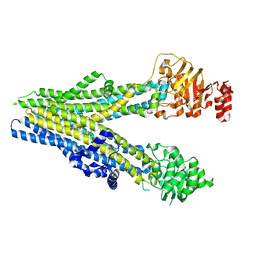 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (in complex with topotecan) | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, ATP-binding cassette sub-family C member 4 | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BWR
 
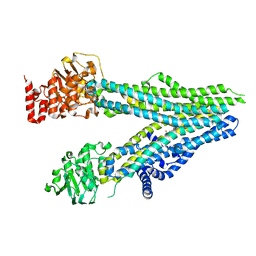 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (in complex with prostaglandin E2) | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, ATP-binding cassette sub-family C member 4 | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BWP
 
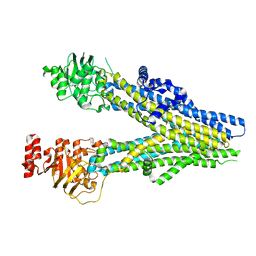 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (in complex with methotrexate) | | Descriptor: | ATP-binding cassette sub-family C member 4, METHOTREXATE | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
6PUY
 
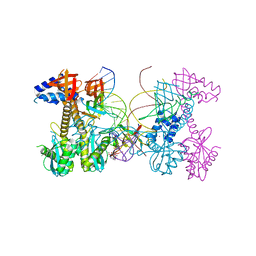 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ426 (compound 4d) | | Descriptor: | 4-amino-N-[(2,4-difluorophenyl)methyl]-1-hydroxy-6-(6-hydroxyhexyl)-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6PW5
 
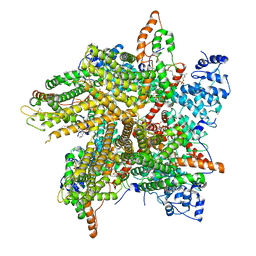 | | Cryo-EM Structure of Thermo-Sensitive TRP Channel TRP1 from the Alga Chlamydomonas reinhardtii in Nanodiscs | | Descriptor: | (2S)-3-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, TRP-like ion channel, ... | | Authors: | McGoldrick, L.L, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-25 | | Last modified: | 2019-12-04 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of the thermo-sensitive TRP channel TRP1 from the alga Chlamydomonas reinhardtii.
Nat Commun, 10, 2019
|
|
6PY5
 
 | |
