7TIE
 
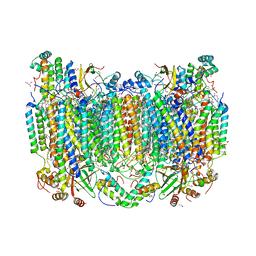 | | Structure of oxidized bovine cytochrome c oxidase at 1.90 Angstrom resolution obtained by synchrotron X-rays | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ishigami, I, Rousseau, D.L, Yeh, S.-R. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Temperature-dependent structural transition following X-ray-induced metal center reduction in oxidized cytochrome c oxidase.
J.Biol.Chem., 298, 2022
|
|
7TIH
 
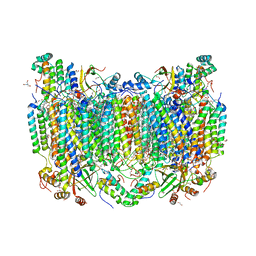 | | Structure of oxidized bovine cytochrome c oxidase with reduced metal centers induced by synchrotron X-ray exposure | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ishigami, I, Rousseau, D.L, Yeh, S.-R, Russi, S, Cohen, A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Temperature-dependent structural transition following X-ray-induced metal center reduction in oxidized cytochrome c oxidase.
J.Biol.Chem., 298, 2022
|
|
6I4K
 
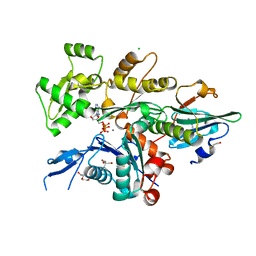 | | Crystal Structure of Plasmodium falciparum actin I (G115A mutant) in the Ca-ATP state | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin-1, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
6I4J
 
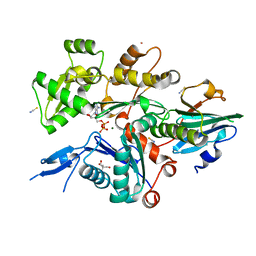 | | Crystal Structure of Plasmodium falciparum actin I (F54Y mutant) in the Mg-ADP state | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Actin-1, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
6I4L
 
 | | Crystal Structure of Plasmodium falciparum actin I (G115A mutant) in the Mg-K-ATP/ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-1, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
1LWF
 
 | | CRYSTAL STRUCTURE OF A MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMQ+M184V: M41L/D67N/K70R/M184V/T215Y) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1FOK
 
 | | STRUCTURE OF RESTRICTION ENDONUCLEASE FOKI BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*TP*GP*AP*CP*TP*AP*GP*CP*GP*TP*TP*AP*TP*CP*AP*T P*CP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*GP*AP*TP*GP*AP*TP*AP*AP*CP*GP*CP*TP*AP*G P*TP*CP*A)-3'), PROTEIN (FOKI RESTRICTION ENDONUCLEASE) | | Authors: | Aggarwal, D.A, Wah, J.A, Hirsch, L.F, Dorner, I, Schildkraut, A.K. | | Deposit date: | 1997-04-18 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the multimodular endonuclease FokI bound to DNA.
Nature, 388, 1997
|
|
3EJH
 
 | | Crystal Structure of the Fibronectin 8-9FnI Domain Pair in Complex with a Type-I Collagen Peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Collagen type-I a1 chain, Fibronectin, ... | | Authors: | Erat, M.C, Lowe, E.D, Campbell, I.D, Vakonakis, I. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and structural analysis of type I collagen sites in complex with fibronectin fragments.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6I4F
 
 | | Crystal Structure of Plasmodium falciparum actin I (A272W mutant) in the Mg-K-ATP/ADP state | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
6I4D
 
 | | Crystal Structure of Plasmodium falciparum actin I in the Mg-K-ATP/ADP state | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
8FEC
 
 | | Structure of J-PKAc chimera complexed with Aplithianine derivative | | Descriptor: | 6-[(6P)-6-(4-bromo-1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7H-purine, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
8ER5
 
 | | Crystal Structure of NlpC/P60 domain from Clostridium innocuum NlpC/P60 domain-containing protein CI_01448. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NlpC/P60 domain-containing protein, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of NlpC/P60 domain from Clostridium innocuum NlpC/P60 domain-containing protein CI_01448.
To Be Published
|
|
8CP6
 
 | |
8EVO
 
 | |
8EVN
 
 | |
5F8W
 
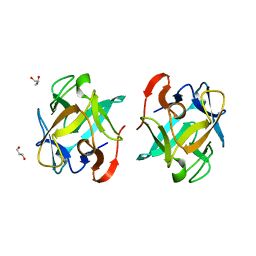 | | Crystal structure of a Crenomytilus grayanus lectin in complex with galactose | | Descriptor: | GLYCEROL, GalNAc/Gal-specific lectin, alpha-D-galactopyranose, ... | | Authors: | Liao, J.-H, Huang, K.-F, Tu, I.-F, Lee, I.-M, Wu, S.-H. | | Deposit date: | 2015-12-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A Multivalent Marine Lectin from Crenomytilus grayanus Possesses Anti-cancer Activity through Recognizing Globotriose Gb3
J.Am.Chem.Soc., 138, 2016
|
|
5OPC
 
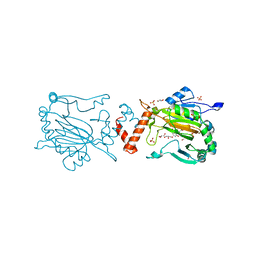 | | Factor Inhibiting HIF (FIH) in complex with zinc and Vadadustat | | Descriptor: | GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Leissing, T.M, Schofield, C.J, Clifton, I.J, Lu, X, Zhang, D. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials.
Chem Sci, 8, 2017
|
|
5M7O
 
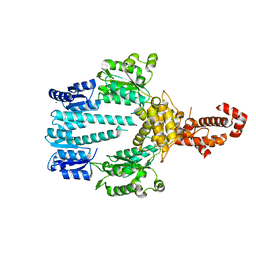 | | Crystal structure of NtrX from Brucella abortus processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | MAGNESIUM ION, Nitrogen assimilation regulatory protein | | Authors: | Cornaciu, I, Fernandez, I, Hoffmann, G, Carrica, M.C, Goldbaum, F.A, Marquez, J.A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-Dimensional Structure of Full-Length NtrX, an Unusual Member of the NtrC Family of Response Regulators.
J. Mol. Biol., 429, 2017
|
|
7QNX
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-55 heavy chain, Beta-55 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
6BGF
 
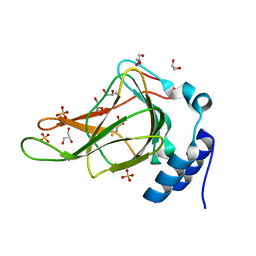 | | Crystal structure of cysteine-bound ferrous form of the crosslinked human cysteine dioxygenase | | Descriptor: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION, ... | | Authors: | Liu, A, Li, J, Shin, I. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Cleavage of a carbon-fluorine bond by an engineered cysteine dioxygenase.
Nat.Chem.Biol., 14, 2018
|
|
8QAA
 
 | |
7T2V
 
 | |
1GD9
 
 | | CRYSTALL STRUCTURE OF PYROCOCCUS PROTEIN-A1 | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Harata, K, Matsui, I, Kuramitsu, S. | | Deposit date: | 2000-09-22 | | Release date: | 2001-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature dependence of the enzyme-substrate recognition mechanism.
J.Biochem., 129, 2001
|
|
8F6R
 
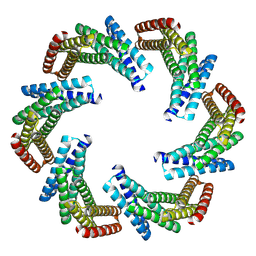 | | CryoEM structure of designed modular protein oligomer C6-79 | | Descriptor: | De novo designed oligomeric protein C6-79 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2022-11-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies.
Cell, 187, 2024
|
|
1RJZ
 
 | | Mhc Class I Natural Mutant H-2Kbm8 Heavy Chain Complexed With beta-2 Microglobulin and Herpies Simplex Virus Mutant Glycoprotein B Peptide | | Descriptor: | Beta-2-microglobulin, Glycoprotein B, H-2 class I histocompatibility antigen, ... | | Authors: | Miley, M.J, Messaoudi, I, Nikolich-Zugich, J, Fremont, D.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Restoration of TCR Recognition of an MHC Allelic Variant by Peptide Secondary Anchor Substitution
J.Exp.Med., 200, 2004
|
|
