8OJ0
 
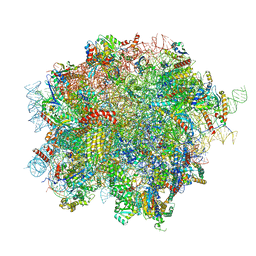 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 2 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
3K2S
 
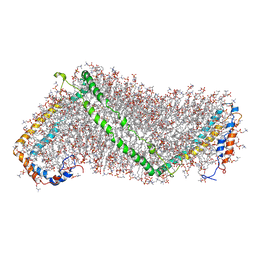 | | Solution structure of double super helix model | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Apolipoprotein A-I, CHOLESTEROL | | Authors: | Wu, Z, Gogonea, V, Lee, X, Wagner, M.A, Li, X.-M, Huang, Y, Undurti, A, May, R.P, Haertlein, M, Moulin, M, Gutsche, I, Zaccai, G, Didonato, J.A, Hazen, L.S. | | Deposit date: | 2009-09-30 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Double superhelix model of high density lipoprotein.
J.Biol.Chem., 284, 2009
|
|
8VDU
 
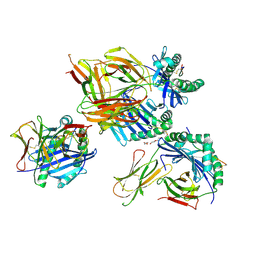 | | Crystal structure of hybrid insulin peptide (InsC8-15-IAPP74-80) bound to HLA-DQ8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Hybrid insulin peptide (HIP; InsC8-15-IAPP74-80), ... | | Authors: | Tran, T.M, Lim, J.J, Loh, T.Y, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2023-12-17 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A structural basis of T cell cross-reactivity to native and spliced self-antigens presented by HLA-DQ8.
J.Biol.Chem., 300, 2024
|
|
7ANY
 
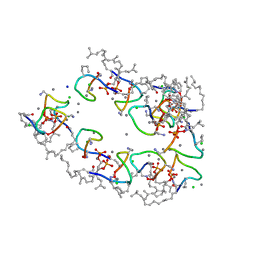 | | Structure of the Laspartomycin C Friulimicin-like mutant in complex with Geranyl phosphate | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zeronian, M.R, Pearce, N.M, Lutz, M, Wood, T.M, Martin, N.I, Janssen, B.J.C. | | Deposit date: | 2020-10-13 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.135 Å) | | Cite: | Mechanistic insights into the C55-P targeting lipopeptide antibiotics revealed by structure-activity studies and high-resolution crystal structures
Chem Sci, 13, 2022
|
|
8QOI
 
 | | Structure of the human 80S ribosome at 1.9 A resolution - the molecular role of chemical modifications and ions in RNA | | Descriptor: | 18S rRNA (1740-MER), 28S rRNA (3773-MER), 40S ribosomal protein S10, ... | | Authors: | Holvec, S, Barchet, C, Frechin, L, Hazemann, I, von Loeffelholz, O, Klaholz, B.P. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | The structure of the human 80S ribosome at 1.9 angstrom resolution reveals the molecular role of chemical modifications and ions in RNA.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7AE6
 
 | | Crystal structure of di-AMPylated HEPN(R102A) toxin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, HEPN toxin | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
4Z5R
 
 | | Rontalizumab Fab bound to Interferon-a2 | | Descriptor: | Interferon alpha-2, SULFATE ION, anti-IFN-a antibody rontalizumab heavy chain modules VH and CH1 (Fab), ... | | Authors: | Eigenbrot, C, Maurer, B, Bosanac, I. | | Deposit date: | 2015-04-02 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of the broadly neutralizing anti-interferon-alpha antibody rontalizumab.
Protein Sci., 24, 2015
|
|
4GHA
 
 | | Crystal structure of Marburg virus VP35 RNA binding domain bound to 12-bp dsRNA | | Descriptor: | Polymerase cofactor VP35, RNA (5'-R(*CP*UP*AP*GP*AP*CP*GP*UP*CP*UP*AP*G)-3') | | Authors: | Bale, S, Jean-Philippe, J, Bornholdt, Z.A, Kimberlin, C.K, Halfmann, P, Zandonatti, M.A, Kunert, J, Kroon, G.J.A, Kawaoka, Y, MacRae, I.J, Wilson, I.A, Saphire, E.O. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Marburg Virus VP35 Can Both Fully Coat the Backbone and Cap the Ends of dsRNA for Interferon Antagonism.
Plos Pathog., 8, 2012
|
|
6MF0
 
 | | Crystal Structure Determination of Human/Porcine Chimera Coagulation Factor VIII | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Smith, I.W, Spiegel, P.C. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.19999886 Å) | | Cite: | The 3.2 angstrom structure of a bioengineered variant of blood coagulation factor VIII indicates two conformations of the C2 domain.
J.Thromb.Haemost., 18, 2020
|
|
6ZXF
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State G | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Ameismeier, M, Zemp, I, van den Heuvel, J, Thoms, M, Berninghausen, O, Kutay, U, Beckmann, R. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for the final steps of human 40S ribosome maturation.
Nature, 587, 2020
|
|
8P3T
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 1 | | Descriptor: | Glutamate receptor 1 flip isoform, Voltage-dependent calcium channel gamma-3 subunit | | Authors: | Zhang, D, Krieger, J, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
6WVT
 
 | | Structural basis of alphaE-catenin - F-actin catch bond behavior | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Xu, X.P, Pokutta, S, Torres, M, Swift, M.F, Hanein, D, Volkmann, N, Weis, W.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of alpha E-catenin-F-actin catch bond behavior.
Elife, 9, 2020
|
|
4RPI
 
 | | Crystal Structure of Nicotinate Mononucleotide Adenylyltransferase from Mycobacterium tuberculosis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, nicotinate-nucleotide adenylyltransferase | | Authors: | Rodionova, I, Zuccola, H, Sorci, L, Aleshin, A.E, Kazanov, M, Sergienko, E, Rubin, E, Locher, C, Osterman, A. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | Mycobacterial nicotinate mononucleotide adenylyltransferase: structure, mechanism, and implications for drug discovery.
J. Biol. Chem., 290, 2015
|
|
8VRB
 
 | | Structure of a synthetic antibody in complex with a class I MHC presenting a hapten-peptide conjugate | | Descriptor: | AMG 510 (bound form), Beta-2-microglobulin, GTPase KRas, ... | | Authors: | Maso, L, Bang, I, Koide, S. | | Deposit date: | 2024-01-20 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Molecular basis for antibody recognition of multiple drug-peptide/MHC complexes.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1WKM
 
 | | THE PRODUCT BOUND FORM OF THE MN(II)LOADED METHIONINE AMINOPEPTIDASE FROM HYPERTHERMOPHILE PYROCOCCUS FURIOSUS | | Descriptor: | MANGANESE (II) ION, METHIONINE, Methionine aminopeptidase | | Authors: | Copik, A.J, Nocek, B.P, Jang, S.B, Swierczek, S.I, Ruebush, S, Meng, L, D'souza, V.M, Peters, J.W, Bennett, B, Holz, R.C. | | Deposit date: | 2004-06-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | EPR and X-ray crystallographic characterization of the product-bound form of the MnII-loaded methionyl aminopeptidase from Pyrococcus furiosus
Biochemistry, 44, 2005
|
|
8F3D
 
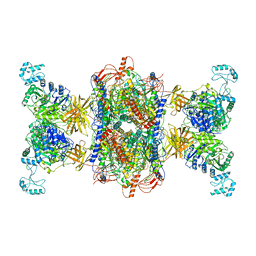 | | 3-methylcrotonyl-CoA carboxylase in filament, beta-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase alpha-subunit, 3-methylcrotonyl-CoA carboxylase beta-subunit, 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-09 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
7B3K
 
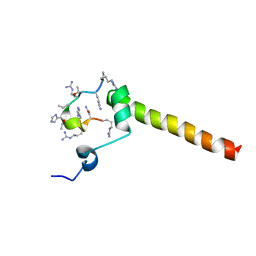 | | Dynamic complex between all-D-enantiomeric peptide D3 with L723P mutant of amyloid precursor protein (APP) 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
5JPM
 
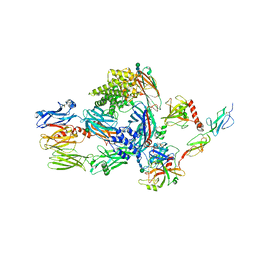 | | Structure of the complex of human complement C4 with MASP-2 rebuilt using iMDFF | | Descriptor: | Complement C4-A, Mannan-binding lectin serine protease 2, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Croll, T.I, Andersen, G.R. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Re-evaluation of low-resolution crystal structures via interactive molecular-dynamics flexible fitting (iMDFF): a case study in complement C4.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5JS4
 
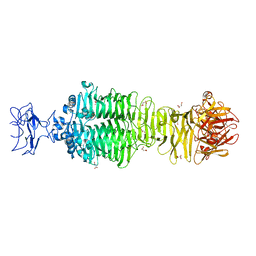 | | Crystal structure of phiAB6 tailspike | | Descriptor: | MALONIC ACID, phiAB6 tailspike | | Authors: | Lee, I.M, Tu, I.F, Huang, K.F, Wu, S.H. | | Deposit date: | 2016-05-07 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural basis for fragmenting the exopolysaccharide of Acinetobacter baumannii by bacteriophage Phi AB6 tailspike protein
Sci Rep, 7, 2017
|
|
1WQJ
 
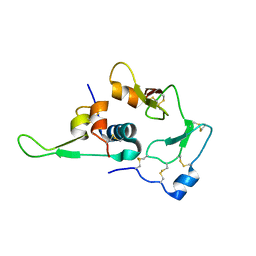 | | Structural Basis for the Regulation of Insulin-Like Growth Factors (IGFs) by IGF Binding Proteins (IGFBPs) | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor binding protein 4 | | Authors: | Siwanowicz, I, Popowicz, G.M, Wisniewska, M, Huber, R, Kuenkele, K.P, Lang, K, Engh, R.A, Holak, T.A. | | Deposit date: | 2004-09-29 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the regulation of insulin-like growth factors by IGF binding proteins
Structure, 13, 2005
|
|
5O9Y
 
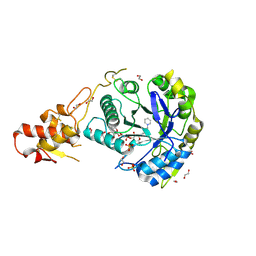 | | Crystal structure of ScGas2 in complex with compound 11 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-3,5-bis(oxidanyl)-6-[4-(3-pyridin-1-ium-1-ylpropyl)-1,2,3-triazol-1-yl]oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2, ... | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
8OOU
 
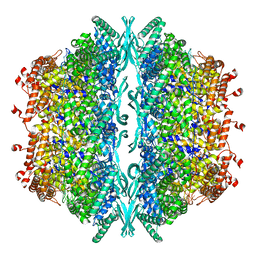 | |
6ZTY
 
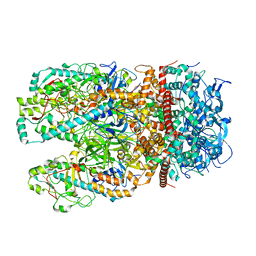 | |
8OP1
 
 | | Subsection of a helical nucleocapsid of the Respiratory Syncytial Virus | | Descriptor: | Nucleoprotein, RNA (5'-R(P*CP*CP*CP*CP*CP*CP*C)-3') | | Authors: | Gonnin, L, Desfosses, A, Eleouet, J.F, Galloux, M, Gutsche, I. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural landscape of the respiratory syncytial virus nucleocapsids.
Nat Commun, 14, 2023
|
|
6T66
 
 | | Crystal structure of the Vibrio cholerae replicative helicase (DnaB) with GDP-AlF4 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Replicative DNA helicase, ... | | Authors: | Legrand, P, Quevillon-Cheruel, S, Li de la Sierra-Gallay, I, Walbott, H. | | Deposit date: | 2019-10-17 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Study of the DnaB:DciA interplay reveals insights into the primary mode of loading of the bacterial replicative helicase.
Nucleic Acids Res., 49, 2021
|
|
