3NDD
 
 | |
5E8E
 
 | | Crystal structure of thrombin bound to an exosite 1-specific IgA Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Baglin, T.P, Langdown, J, Frasson, R, Huntington, J.A. | | Deposit date: | 2015-10-14 | | Release date: | 2015-10-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and characterization of an antibody directed against exosite I of thrombin.
J.Thromb.Haemost., 14, 2016
|
|
4CH2
 
 | | Low-salt crystal structure of a thrombin-GpIbalpha peptide complex | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, PLATELET GLYCOPROTEIN IB ALPHA CHAIN, ... | | Authors: | Lechtenberg, B.C, Freund, S.M.V, Huntington, J.A. | | Deposit date: | 2013-11-28 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gpibalpha Interacts Exclusively with Exosite II of Thrombin
J.Mol.Biol., 426, 2014
|
|
4RBM
 
 | | Porphyromonas gingivalis gingipain K (Kgp) catalytic and immunoglobulin superfamily-like domains | | Descriptor: | (3S)-3,7-diaminoheptan-2-one, ACETATE ION, AZIDE ION, ... | | Authors: | de Diego, I, Veillard, F, Sztukowska, M.N, Guevara, T, Potempa, B, Pomowski, A, Huntington, J.A, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Mechanism of Cysteine Peptidase Gingipain K (Kgp), a Major Virulence Factor of Porphyromonas gingivalis in Periodontitis.
J.Biol.Chem., 289, 2014
|
|
5MUN
 
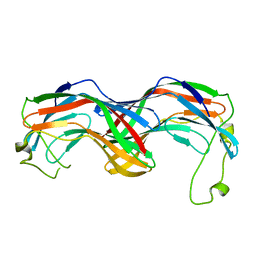 | | Structural insight into zymogenic latency of gingipain K from Porphyromonas gingivalis. | | Descriptor: | AZIDE ION, Lys-gingipain W83 | | Authors: | Pomowski, A, Uson, I, Nowakovska, Z, Veillard, F, Sztukowska, M.N, Guevara, T, Goulas, T, Mizgalska, D, Nowak, M, Potempa, B, Huntington, J.A, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights unravel the zymogenic mechanism of the virulence factor gingipain K from Porphyromonas gingivalis, a causative agent of gum disease from the human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
1JB2
 
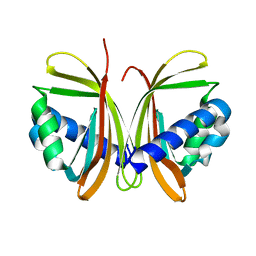 | | CRYSTAL STRUCTURE OF NTF2 M84E MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Chaillan-Huntington, C, Butler, P.J, Huntington, J.A, Akin, D, Feldherr, C, Stewart, M. | | Deposit date: | 2001-06-01 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | NTF2 monomer-dimer equilibrium.
J.Mol.Biol., 314, 2001
|
|
1JB4
 
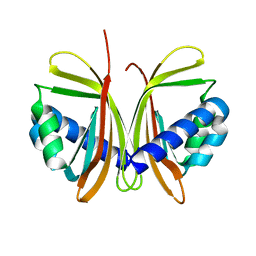 | | CRYSTAL STRUCTURE OF NTF2 M102E MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Chaillan-Huntington, C, Butler, P.J, Huntington, J.A, Akin, D, Feldherr, C, Stewart, M. | | Deposit date: | 2001-06-01 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | NTF2 monomer-dimer equilibrium.
J.Mol.Biol., 314, 2001
|
|
1JB5
 
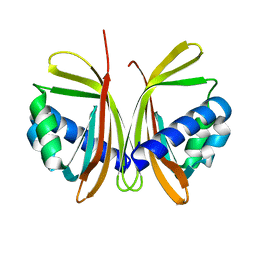 | | CRYSTAL STRUCTURE OF NTF2 M118E MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Chaillan-Huntington, C, Butler, P.J, Huntington, J.A, Akin, D, Feldherr, C, Stewart, M. | | Deposit date: | 2001-06-01 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NTF2 monomer-dimer equilibrium.
J.Mol.Biol., 314, 2001
|
|
1DZH
 
 | | P14-FLUORESCEIN-N135Q-S380C-ANTITHROMBIN-III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, ... | | Authors: | Mccoy, A.J, Huntington, J.A, Carrell, R.W. | | Deposit date: | 2000-02-28 | | Release date: | 2000-05-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Conformational Activation of Antithrombin. A 2. 85-A Structure of a Fluorescein Derivative Reveals an Electrostatic Link between the Hinge and Heparin Binding Regions.
J.Biol.Chem., 275, 2000
|
|
1DZG
 
 | | N135Q-S380C-ANTITHROMBIN-III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, ... | | Authors: | McCoy, A.J, Huntington, J.A, Carrell, R.W. | | Deposit date: | 2000-02-28 | | Release date: | 2000-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Conformational Activation of Antithrombin. A 2. 85-A Structure of a Fluorescein Derivative Reveals an Electrostatic Link between the Hinge and Heparin Binding Regions.
J.Biol.Chem., 275, 2000
|
|
1BR8
 
 | | IMPLICATIONS FOR FUNCTION AND THERAPY OF A 2.9A STRUCTURE OF BINARY-COMPLEXED ANTITHROMBIN | | Descriptor: | PROTEIN (ANTITHROMBIN-III), PROTEIN (PEPTIDE) | | Authors: | Skinner, R, Chang, W.S.W, Jin, L, Pei, X.Y, Huntington, J.A, Abrahams, J.P, Carrell, R.W, Lomas, D.A. | | Deposit date: | 1998-08-26 | | Release date: | 1998-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Implications for function and therapy of a 2.9 A structure of binary-complexed antithrombin.
J.Mol.Biol., 283, 1998
|
|
1OYH
 
 | | Crystal Structure of P13 Alanine Variant of Antithrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III, ... | | Authors: | Johnson, D.J.D, Huntington, J.A. | | Deposit date: | 2003-04-04 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The influence of hinge region residue Glu-381 on antithrombin allostery and metastability
J.Biol.Chem., 279, 2004
|
|
1R1L
 
 | | Structure of dimeric antithrombin complexed with a P14-P9 reactive loop peptide and an exogenous tripeptide (formyl-norleucine-LF) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin P14-P9 peptide, Antithrombin-III, ... | | Authors: | Zhou, A, Huntington, J.A, Lomas, D.A, Stein, P.E, Carrell, R.W. | | Deposit date: | 2003-09-24 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Serpins and the design of peptides to block intermolecular beta-linkages
To be Published
|
|
1RD3
 
 | | 2.5A Structure of Anticoagulant Thrombin Variant E217K | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Carter, W.J, Myles, T, Leung, L.L, Huntington, J.A. | | Deposit date: | 2003-11-05 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Anticoagulant Thrombin Variant E217K Provides Insights into Thrombin Allostery
J.Biol.Chem., 279, 2004
|
|
1T1F
 
 | | Crystal Structure of Native Antithrombin in its Monomeric Form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III, GLYCEROL, ... | | Authors: | Johnson, D.J.D, Huntington, J.A. | | Deposit date: | 2004-04-16 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of monomeric native antithrombin reveals a novel reactive center loop conformation
J.Biol.Chem., 281, 2006
|
|
1TB6
 
 | | 2.5A Crystal Structure of the Antithrombin-Thrombin-Heparin Ternary Complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfonato-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-alpha-L-idopyranuronic acid-(1-4)-methyl 3-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Johnson, D.J, Esmon, C.T, Huntington, J.A. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the antithrombin-thrombin-heparin ternary complex reveals the antithrombotic mechanism of heparin.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2AFQ
 
 | |
2BEH
 
 | | Crystal structure of antithrombin variant S137A/V317C/T401C with plasma latent antithrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III, ... | | Authors: | Johnson, D.J, Luis, S.A, Huntington, J.A. | | Deposit date: | 2005-10-24 | | Release date: | 2005-11-01 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of monomeric native antithrombin reveals a novel reactive center loop conformation.
J.Biol.Chem., 281, 2006
|
|
4BXS
 
 | | Crystal Structure of the Prothrombinase Complex from the Venom of Pseudonaja Textilis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lechtenberg, B.C, Murray-Rust, T.A, Johnson, D.J.D, Adams, T.E, Krishnaswamy, S, Camire, R.M, Huntington, J.A. | | Deposit date: | 2013-07-15 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Crystal Structure of the Prothrombinase Complex from the Venom of Pseudonaja Textilis.
Blood, 122, 2013
|
|
2B5T
 
 | | 2.1 Angstrom structure of a nonproductive complex between antithrombin, synthetic heparin mimetic SR123781 and two S195A thrombin molecules | | Descriptor: | 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfonato-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-alpha-L-idopyranuronic acid-(1-4)-methyl 3-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Johnson, D.J, Li, W, Luis, S.A, Carrell, R.W, Huntington, J.A. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of monomeric native antithrombin reveals a novel reactive center loop conformation.
J.Biol.Chem., 281, 2006
|
|
2B4X
 
 | |
4BXW
 
 | | Crystal Structure of the Prothrombinase Complex from the Venom of Pseudonaja Textilis | | Descriptor: | COAGULATION FACTOR V, FACTOR XA, GLYCEROL, ... | | Authors: | Lechtenberg, B.C, Murray-Rust, T.A, Johnson, D.J.D, Adams, T.E, Krishnaswamy, S, Camire, R.M, Huntington, J.A. | | Deposit date: | 2013-07-16 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structure of the Prothrombinase Complex from the Venom of Pseudonaja Textilis.
Blood, 122, 2013
|
|
4CH8
 
 | | High-salt crystal structure of a thrombin-GpIbalpha peptide complex | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, PLATELET GLYCOPROTEIN IB ALPHA CHAIN, ... | | Authors: | Lechtenberg, B.C, Freund, S.M.V, Huntington, J.A. | | Deposit date: | 2013-11-29 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Gpibalpha Interacts Exclusively with Exosite II of Thrombin
J.Mol.Biol., 426, 2014
|
|
3DY0
 
 | | Crystal Structure of Cleaved PCI Bound to Heparin | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, C-terminus Plasma serine protease inhibitor, GLYCEROL, ... | | Authors: | Li, W, Huntington, J.A. | | Deposit date: | 2008-07-25 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The heparin binding site of protein C inhibitor is protease-dependent.
J.Biol.Chem., 283, 2008
|
|
3B9F
 
 | | 1.6 A structure of the PCI-thrombin-heparin complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, GLYCEROL, Plasma serine protease inhibitor, ... | | Authors: | Li, W, Adams, T.E, Huntington, J.A. | | Deposit date: | 2007-11-05 | | Release date: | 2008-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of thrombin recognition by protein C inhibitor revealed by the 1.6-A structure of the heparin-bridged complex.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
