8USM
 
 | | FmlH Lectin Domain UTI89 - AM4085 | | Descriptor: | 4'-fluoro-6-(trifluoromethyl)[1,1'-biphenyl]-2-yl 2-acetamido-2-deoxy-beta-D-galactopyranoside, Fimbrial protein (Fragment) | | Authors: | Tamadonfar, K.O, Pinkner, J.P, Maddirala, A.R, Janetka, J.W, Hultgren, S.J. | | Deposit date: | 2023-10-27 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of Orally Bioavailable FmlH Lectin Antagonists as Treatment for Urinary Tract Infections.
J.Med.Chem., 67, 2024
|
|
6MAW
 
 | | F9 Pilus Adhesin FmlH Lectin Domain from E. coli UTI89 in Complex with Galactoside N-[(2S,3R,4R,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)-2-{[S-methyl-6-(trifluoromethyl)-[1,1'-biphenyl]-3'-yl]oxy}oxan-3-yl]acetamide | | Descriptor: | Fimbrial adhesin FmlD, N-[2'-{[2-(acetylamino)-2-deoxy-beta-D-galactopyranosyl]oxy}-6'-(trifluoromethyl)[1,1'-biphenyl]-3-yl]methanesulfonamide | | Authors: | Klein, R.D, Hultgren, S.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biphenyl Gal and GalNAc FmlH Lectin Antagonists of Uropathogenic E. coli (UPEC): Optimization through Iterative Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
1QUN
 
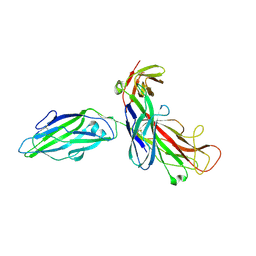 | | X-RAY STRUCTURE OF THE FIMC-FIMH CHAPERONE ADHESIN COMPLEX FROM UROPATHOGENIC E.COLI | | Descriptor: | MANNOSE-SPECIFIC ADHESIN FIMH, PAPD-LIKE CHAPERONE FIMC | | Authors: | Choudhury, D, Thompson, A, Stojanoff, V, Langerman, S, Pinkner, J, Hultgren, S.J, Knight, S. | | Deposit date: | 1999-07-01 | | Release date: | 1999-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli.
Science, 285, 1999
|
|
6MAP
 
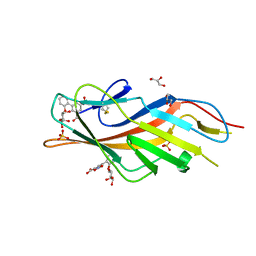 | | F9 Pilus Adhesin FmlH Lectin Domain from E. coli UTI89 in Complex with Galactoside 5-nitro-2'-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-[1,1'-biphenyl]-3-carboxylic acid | | Descriptor: | 2'-(beta-D-galactopyranosyloxy)-5-nitro[1,1'-biphenyl]-3-carboxylic acid, Fimbrial adhesin FmlD, GLYCEROL, ... | | Authors: | Klein, R.D, Hultgren, S.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Biphenyl Gal and GalNAc FmlH Lectin Antagonists of Uropathogenic E. coli (UPEC): Optimization through Iterative Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
9AT9
 
 | |
4UV3
 
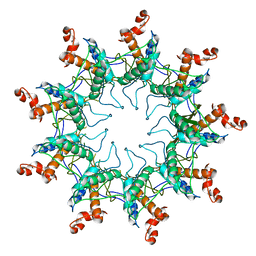 | | Structure of the curli transport lipoprotein CsgG in its membrane- bound conformation | | Descriptor: | CURLI PRODUCTION ASSEMBLY/TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
4UV2
 
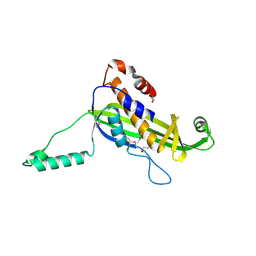 | | Structure of the curli transport lipoprotein CsgG in a non-lipidated, pre-pore conformation | | Descriptor: | CURLI PRODUCTION TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
3L48
 
 | |
8DKA
 
 | |
3ME0
 
 | | Structure of the E. coli chaperone PAPD in complex with the pilin domain of the PapGII adhesin | | Descriptor: | Chaperone protein papD, PapG protein | | Authors: | Ford, B.A, Elam, J.S, Dodson, K.W, Pinkner, J.S, Hultgren, S.J. | | Deposit date: | 2010-03-31 | | Release date: | 2011-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the E. Coli Chaperone PapD in Complex with the Pilin Domain of the PapGII Adhesin
To be Published
|
|
3MCY
 
 | |
5JR4
 
 | |
5JQI
 
 | |
5LNG
 
 | | Lectin domain of E. coli F9 pilus adhesin FmlH | | Descriptor: | Putative Fml fimbrial adhesin FmlD, SULFATE ION | | Authors: | Ruer, S, Conover, M.S, Kalas, V, Taganna, J, De Greve, H, Pinkner, J.S, Dodson, K.W, Hultgren, S.J, Remaut, H. | | Deposit date: | 2016-08-04 | | Release date: | 2016-10-05 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Inflammation-Induced Adhesin-Receptor Interaction Provides a Fitness Advantage to Uropathogenic E. coli during Chronic Infection.
Cell Host Microbe, 20, 2016
|
|
5LNE
 
 | | E. coli F9 pilus adhesin FmlH bound to the Thomsen-Friedenreich (TF) antigen | | Descriptor: | NICKEL (II) ION, Putative Fml fimbrial adhesin FmlD, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Ruer, S, Conover, M.S, Kalas, V, Taganna, J, De Greve, H, Pinkner, J.S, Dodson, K.W, Hultgren, S.J, Remaut, H. | | Deposit date: | 2016-08-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inflammation-Induced Adhesin-Receptor Interaction Provides a Fitness Advantage to Uropathogenic E. coli during Chronic Infection.
Cell Host Microbe, 20, 2016
|
|
8DEZ
 
 | | Abp2D Receptor Binding Domain ACICU | | Descriptor: | Abp2D Receptor Binding Domain, CITRATE ANION, SULFATE ION | | Authors: | Tamadonfar, K.O, Pinkner, J.P, Dodson, K.W, Kalas, V, Hultgren, S.J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structure-function correlates of fibrinogen binding by Acinetobacter adhesins critical in catheter-associated urinary tract infections.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DF0
 
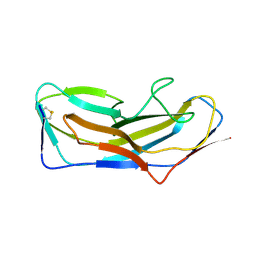 | | Abp1D receptor binding domain | | Descriptor: | Abp1D Receptor Binding Domain | | Authors: | Tamadonfar, K.O, Pinkner, J.S, Dodson, K.W, Hultgren, S.J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-function correlates of fibrinogen binding by Acinetobacter adhesins critical in catheter-associated urinary tract infections.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6MAQ
 
 | | F9 Pilus Adhesin FmlH Lectin Domain from E. coli UTI89 in Complex with Galactoside 2'-{[(2S,3R,4R,5R,6R)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-5-nitro-[1,1'-biphenyl]-3-carboxylic acid | | Descriptor: | 2'-{[2-(acetylamino)-2-deoxy-beta-D-galactopyranosyl]oxy}-5-nitro[1,1'-biphenyl]-3-carboxylic acid, Fimbrial adhesin FmlD, GLYCEROL | | Authors: | Klein, R.D, Hultgren, S.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Biphenyl Gal and GalNAc FmlH Lectin Antagonists of Uropathogenic E. coli (UPEC): Optimization through Iterative Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
3PNT
 
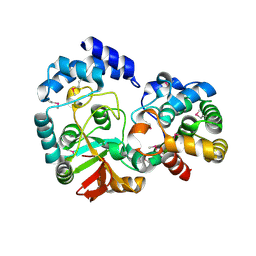 | | Crystal Structure of the Streptococcus pyogenes NAD+ glycohydrolase SPN in complex with IFS, the Immunity Factor for SPN | | Descriptor: | Immunity factor for SPN, NAD+-glycohydrolase | | Authors: | Smith, C.L, Stine Elam, J, Ellenberger, T, Ghosh, J, Pinkner, J.S, Hultgren, S.J, Caparon, M.G. | | Deposit date: | 2010-11-19 | | Release date: | 2011-03-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Streptococcus pyogenes Immunity to Its NAD(+) Glycohydrolase Toxin.
Structure, 19, 2011
|
|
2XG5
 
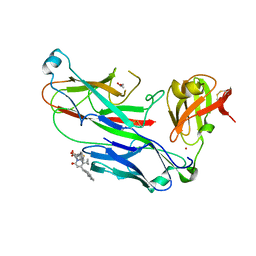 | | E. coli P pilus chaperone-subunit complex PapD-PapH bound to pilus biogenesis inhibitor, pilicide 5d | | Descriptor: | (2R)-2-[5-CYCLOPROPYL-6-(HYDROXYSULFANYL)-4-(NAPHTHALEN-1-YLMETHYL)-2-OXOPYRIDIN-1(2H)-YL]-3-PHENYLPROPANOIC ACID, (2R,3R)-8-CYCLOPROPYL-7-(NAPHTHALEN-1-YLMETHYL)-5-OXO-2-PHENYL-2,3-DIHYDRO-5H-[1,3]THIAZOLO[3,2-A]PYRIDINE-3-CARBOXYLIC ACID, CHAPERONE PROTEIN PAPD, ... | | Authors: | Remaut, H, Phan, G, Buelens, F, Chorell, E, Pinkner, J.S, Edvinsson, S, Almqvist, F, Hultgren, S.J, Waksman, G. | | Deposit date: | 2010-05-30 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of C-2 Substituted Thiazolo and Dihydrothiazolo Ring-Fused 2-Pyridones: Pilicides with Increased Antivirulence Activity.
J.Med.Chem., 53, 2010
|
|
2XG4
 
 | | E. coli P pilus chaperone-subunit complex PapD-PapH bound to pilus biogenesis inhibitor, pilicide 2c | | Descriptor: | (3R)-8-CYCLOPROPYL-6-(MORPHOLIN-4-YLMETHYL)-7-(1-NAPHTHYLMETHYL)-5-OXO-2,3-DIHYDRO-5H-[1,3]THIAZOLO[3,2-A]PYRIDINE-3-CARBOXYLIC ACID, CHAPERONE PROTEIN PAPD, COBALT (II) ION, ... | | Authors: | Remaut, H, Phan, G, Buelens, F, Chorell, E, Pinkner, J.S, Edvinsson, S, Almqvist, F, Hultgren, S.J, Waksman, G. | | Deposit date: | 2010-05-30 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Synthesis of C-2 Substituted Thiazolo and Dihydrothiazolo Ring-Fused 2-Pyridones: Pilicides with Increased Antivirulence Activity.
J.Med.Chem., 53, 2010
|
|
5F2F
 
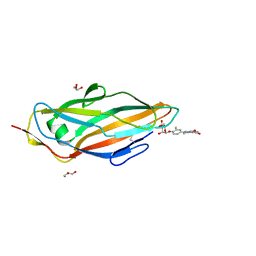 | | Crystal structure of para-biphenyl-2-methyl-3', 5' di-methyl amide mannoside bound to FimH lectin domain | | Descriptor: | 5-[4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-methyl-phenyl]-~{N}1,~{N}3-dimethyl-benzene-1,3-dicarboxamide, GLYCEROL, Protein FimH | | Authors: | Kalas, V, Hultgren, S.J. | | Deposit date: | 2015-12-01 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Antivirulence Isoquinolone Mannosides: Optimization of the Biaryl Aglycone for FimH Lectin Binding Affinity and Efficacy in the Treatment of Chronic UTI.
Chemmedchem, 11, 2016
|
|
5F3F
 
 | | Crystal structure of para-biphenyl-2-methyl-3'-methyl amide mannoside bound to FimH lectin domain | | Descriptor: | 3-[4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-methyl-phenyl]-~{N}-methyl-benzamide, Protein FimH | | Authors: | Klein, R.D, Hultgren, S.J. | | Deposit date: | 2015-12-02 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Antivirulence C-Mannosides as Antibiotic-Sparing, Oral Therapeutics for Urinary Tract Infections.
J.Med.Chem., 59, 2016
|
|
1TR7
 
 | | FimH adhesin receptor binding domain from uropathogenic E. coli | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, FimH protein, ... | | Authors: | Bouckaert, J, Berglund, J, Schembri, M, De Genst, E, Cools, L, Wuhrer, M, Hung, C.S, Pinkner, J, Slattegard, R, Zavialov, A, Choudhury, D, Langermann, S, Hultgren, S.J, Wyns, L, Klemm, P, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-06-21 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin
Mol.Microbiol., 55, 2005
|
|
2WMP
 
 | | Structure of the E. coli chaperone PapD in complex with the pilin domain of the PapGII adhesin | | Descriptor: | CHAPERONE PROTEIN PAPD, PAPG PROTEIN | | Authors: | Ford, B.A, Verger, D, Elam, J.S, Dodson, K.W, Pinkner, J.S, Hultgren, S.J. | | Deposit date: | 2009-07-02 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Papd-Papgii Pilin Complex Reveals an Open and Flexible P5 Pocket.
J.Bacteriol., 194, 2012
|
|
