6QFY
 
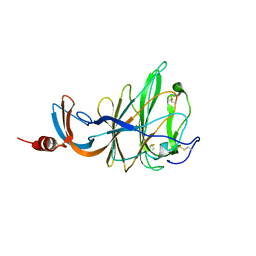 | | CRYSTAL STRUCTURE OF PORCINE HEMAGGLUTINATING ENCEPHALOMYELITIS VIRUS SPIKE PROTEIN LECTIN DOMAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Huizinga, E.G, Bakkers, M, Lang, Y. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1ATZ
 
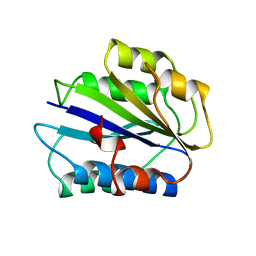 | | HUMAN VON WILLEBRAND FACTOR A3 DOMAIN | | Descriptor: | VON WILLEBRAND FACTOR | | Authors: | Huizinga, E.G, Gros, P. | | Deposit date: | 1997-08-15 | | Release date: | 1998-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the A3 domain of human von Willebrand factor: implications for collagen binding.
Structure, 5, 1997
|
|
1I8N
 
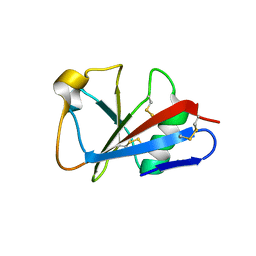 | | CRYSTAL STRUCTURE OF LEECH ANTI-PLATELET PROTEIN | | Descriptor: | ANTI-PLATELET PROTEIN, PROPIONAMIDE | | Authors: | Huizinga, E.G, Schouten, A, Connolly, T.M, Kroon, J, Sixma, J.J, Gros, P. | | Deposit date: | 2001-03-15 | | Release date: | 2001-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of leech anti-platelet protein, an inhibitor of haemostasis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1MAF
 
 | |
1MAE
 
 | |
2MAD
 
 | |
1M10
 
 | | Crystal structure of the complex of Glycoprotein Ib alpha and the von Willebrand Factor A1 Domain | | Descriptor: | Glycoprotein Ib alpha, von Willebrand Factor | | Authors: | Huizinga, E.G, Tsuji, S, Romijn, R.A.P, Schiphorst, M.E, de Groot, P.G, Sixma, J.J, Gros, P. | | Deposit date: | 2002-06-16 | | Release date: | 2002-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain.
Science, 297, 2002
|
|
1M0Z
 
 | | Crystal Structure of the von Willebrand Factor Binding Domain of Glycoprotein Ib alpha | | Descriptor: | Glycoprotein Ib alpha | | Authors: | Huizinga, E.G, Tsuji, S, Romijn, R.A.P, Schiphorst, M.E, de Groot, P.G, Sixma, J.J, Gros, P. | | Deposit date: | 2002-06-16 | | Release date: | 2002-08-28 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain.
Science, 297, 2002
|
|
1FE8
 
 | | CRYSTAL STRUCTURE OF THE VON WILLEBRAND FACTOR A3 DOMAIN IN COMPLEX WITH A FAB FRAGMENT OF IGG RU5 THAT INHIBITS COLLAGEN BINDING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, IMMUNOGLOBULIN IGG RU5, ... | | Authors: | Bouma, B, Huizinga, E.G, Schiphorst, M.E, Sixma, J.J, Kroon, J, Gros, P. | | Deposit date: | 2000-07-21 | | Release date: | 2001-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of the collagen-binding site of the von Willebrand factor A3-domain.
J.Biol.Chem., 276, 2001
|
|
3KGR
 
 | |
3ZQK
 
 | | Von Willebrand Factor A2 domain with calcium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Jakobi, A.J, Huizinga, E.G. | | Deposit date: | 2011-06-09 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Calcium Modulates Force Sensing by the Von Willebrand Factor A2 Domain.
Nat.Commun., 2, 2011
|
|
5JIF
 
 | | Crystal structure of mouse hepatitis virus strain DVIM Hemagglutinin-Esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zeng, Q.H, Bakkers, M.J.G, Feitsma, L.J, de Groot, R.J, Huizinga, E.G. | | Deposit date: | 2016-04-22 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JIL
 
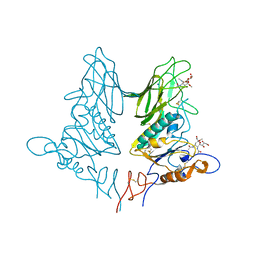 | | Crystal structure of rat coronavirus strain New-Jersey Hemagglutinin-Esterase in complex with 4N-acetyl sialic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Bakkers, M.J.G, Feitsma, L.J, de Groot, R.J, Huizinga, E.G. | | Deposit date: | 2016-04-22 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5N11
 
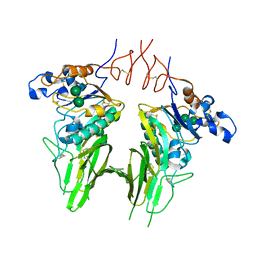 | | Crystal structure of Human beta1-coronavirus OC43 NL/A/2005 Hemagglutinin-Esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Bakkers, M.J.G, Feitsma, L.J, de Groot, R.J, Huizinga, E.G. | | Deposit date: | 2017-02-04 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Betacoronavirus Adaptation to Humans Involved Progressive Loss of Hemagglutinin-Esterase Lectin Activity.
Cell Host Microbe, 21, 2017
|
|
2ADF
 
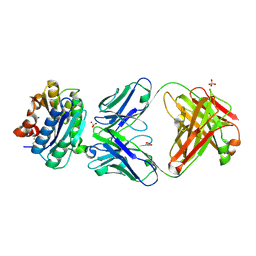 | | Crystal Structure and Paratope Determination of 82D6A3, an Antithrombotic Antibody Directed Against the von Willebrand factor A3-Domain | | Descriptor: | 82D6A3 IgG, ACETIC ACID, GLYCEROL, ... | | Authors: | Staelens, S, Hadders, M.A, Vauterin, S, Platteau, C, Vanhoorelbeke, K, Huizinga, E.G, Deckmyn, H. | | Deposit date: | 2005-07-20 | | Release date: | 2005-12-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Paratope determination of the antithrombotic antibody 82D6A3 based on the crystal structure of its complex with the von Willebrand factor A3-domain
J.Biol.Chem., 281, 2006
|
|
6GRQ
 
 | |
6GRT
 
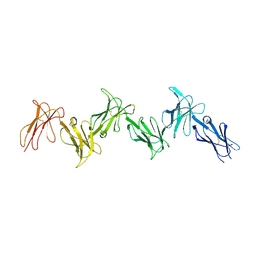 | | Paired immunoglobulin-like receptor B (PirB) or Leukocyte immunoglobulin-like receptor subfamily B member 3 (LILRB3) full extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Paired immunoglobulin-like receptor B, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vlieg, H.C, Huizinga, E.G, Janssen, B.J.C. | | Deposit date: | 2018-06-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.504 Å) | | Cite: | Structure and flexibility of the extracellular region of the PirB receptor.
J.Biol.Chem., 294, 2019
|
|
6GRS
 
 | |
2A74
 
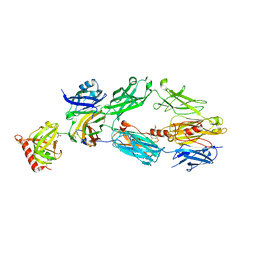 | | Human Complement Component C3c | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Complement Component C3c, GLYCEROL, ... | | Authors: | Janssen, B.J.C, Huizinga, E.G, Raaijmakers, H.C.A, Roos, A, Daha, M.R, Nilsson-Ekdahl, K, Nilsson, B, Gros, P. | | Deposit date: | 2005-07-04 | | Release date: | 2005-09-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of complement component C3 provide insights into the function and evolution of immunity.
Nature, 437, 2005
|
|
2A73
 
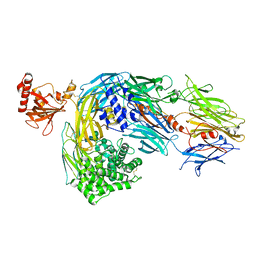 | | Human Complement Component C3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Janssen, B.J.C, Huizinga, E.G, Raaijmakers, H.C.A, Roos, A, Daha, M.R, Nilsson-Ekdahl, K, Nilsson, B, Gros, P. | | Deposit date: | 2005-07-04 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of complement component C3 provide insights into the function and evolution of immunity.
Nature, 437, 2005
|
|
4ZXN
 
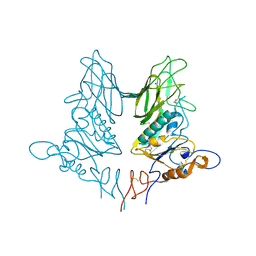 | | Crystal structure of rat coronavirus strain New-Jersey Hemagglutinin-Esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HE protein, SODIUM ION, ... | | Authors: | Bakkers, M.J.G, Feitsma, L.J, Huizinga, E.G, de Groot, R.J. | | Deposit date: | 2015-05-20 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of murine coronavirus hemagglutinin-esterases reveal structural basis for esterase substrate specificity
To Be Published
|
|
5AJ2
 
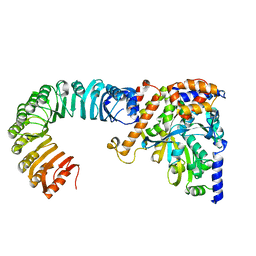 | | Cryo electron tomography of the Naip5-Nlrc4 inflammasome | | Descriptor: | NLR FAMILY CARD DOMAIN-CONTAINING PROTEIN 4 | | Authors: | Diebolder, C.A, Halff, E.F, Koster, A.J, Huizinga, E.G, Koning, R.I. | | Deposit date: | 2015-02-20 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (40 Å) | | Cite: | Cryoelectron Tomography of the Naip5/Nlrc4 Inflammasome: Implications for Nlr Activation.
Structure, 23, 2015
|
|
5D3I
 
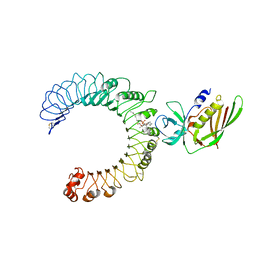 | | Crystal structure of the SSL3-TLR2 complex | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feitsma, L.J, Huizinga, E.G. | | Deposit date: | 2015-08-06 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for inhibition of TLR2 by staphylococcal superantigen-like protein 3 (SSL3).
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5D3D
 
 | |
2I6S
 
 | | Complement component C2a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C2a fragment, ... | | Authors: | Milder, F.J, Raaijmakers, H.C.A, Vandeputte, D.A.A, Schouten, A, Huizinga, E.G, Romijn, R.A, Hemrika, W, Roos, A, Daha, M.R, Gros, P. | | Deposit date: | 2006-08-29 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of complement component c2a: implications for convertase formation and substrate binding.
Structure, 14, 2006
|
|
