1GTJ
 
 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Ala-Phe-cho | | 分子名称: | ALDEHYDE INHIBITOR, CALCIUM ION, KUMAMOLYSIN, ... | | 著者 | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | 登録日 | 2002-01-15 | | 公開日 | 2002-06-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1GTG
 
 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolysin (kscp) | | 分子名称: | CALCIUM ION, KUMAMOLYSIN | | 著者 | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | 登録日 | 2002-01-15 | | 公開日 | 2002-06-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1GV3
 
 | | The 2.0 Angstrom resolution structure of the catalytic portion of a cyanobacterial membrane-bound manganese superoxide dismutase | | 分子名称: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | 著者 | Atzenhofer, W, Regelsberger, G, Jacob, U, Huber, R, Peschek, G.A, Obinger, C. | | 登録日 | 2002-02-05 | | 公開日 | 2002-08-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The 2.0A Resolution Structure of the Catalytic Portion of a Cyanobacterial Membrane-Bound Manganese Superoxide Dismutase
J.Mol.Biol., 321, 2002
|
|
1HOE
 
 | |
6TRX
 
 | | Crystal structure of DPP8 in complex with 1G244 | | 分子名称: | (2~{S})-2-azanyl-4-[4-[bis(4-fluorophenyl)methyl]piperazin-1-yl]-1-(1,3-dihydroisoindol-2-yl)butane-1,4-dione, Dipeptidyl peptidase 8, PHOSPHATE ION, ... | | 著者 | Ross, B.H, Huber, R. | | 登録日 | 2019-12-19 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Aerosol-based ligand soaking of reservoir-free protein crystals.
J.Appl.Crystallogr., 54, 2021
|
|
1QRB
 
 | | PLASTICITY AND STERIC STRAIN IN A PARALLEL BETA-HELIX: RATIONAL MUTATIONS IN P22 TAILSPIKE PROTEIN | | 分子名称: | PROTEIN (TAILSPIKE-PROTEIN) | | 著者 | Schuler, B, Furst, F, Osterroth, F, Steinbacher, S, Huber, R, Seckler, R. | | 登録日 | 1999-06-12 | | 公開日 | 2000-04-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Plasticity and steric strain in a parallel beta-helix: rational mutations in the P22 tailspike protein.
Proteins, 39, 2000
|
|
1QWJ
 
 | | The Crystal Structure of Murine CMP-5-N-Acetylneuraminic Acid Synthetase | | 分子名称: | CYTIDINE-5'-MONOPHOSPHATE-5-N-ACETYLNEURAMINIC ACID, cytidine monophospho-N-acetylneuraminic acid synthetase | | 著者 | Krapp, S, Muenster-Kuehnel, A.K, Kaiser, J.T, Huber, R, Tiralongo, J, Gerardy-Schahn, R, Jacob, U. | | 登録日 | 2003-09-02 | | 公開日 | 2003-12-09 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The Crystal Structure of Murine CMP-5-N-acetylneuraminic Acid Synthetase
J.Mol.Biol., 334, 2003
|
|
6TRW
 
 | |
1QDB
 
 | | CYTOCHROME C NITRITE REDUCTASE | | 分子名称: | CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, HEME C, ... | | 著者 | Einsle, O, Messerschmidt, A, Stach, P, Huber, R, Kroneck, P.M.H. | | 登録日 | 1999-05-19 | | 公開日 | 1999-08-18 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of cytochrome c nitrite reductase.
Nature, 400, 1999
|
|
6T41
 
 | | CDK8/Cyclin C in complex with N-(4-chlorobenzyl)isoquinolin-4-amine | | 分子名称: | 1,2-ETHANEDIOL, Cyclin-C, Cyclin-dependent kinase 8, ... | | 著者 | Schneider, E.V, Maskos, K, Huber, R, Kuhn, C.-D. | | 登録日 | 2019-10-11 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | A precisely positioned MED12 activation helix stimulates CDK8 kinase activity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7ZXS
 
 | | Crystal structure of DPP9 in complex with a 4-oxo-b-lactam based inhibitor, A295 | | 分子名称: | 1,2-ETHANEDIOL, 2-ethyl-2-methanoyl-~{N}-[3-[[4-(quinolin-8-ylmethyl)piperazin-1-yl]methyl]phenyl]butanamide, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Ross, B, Huber, R. | | 登録日 | 2022-05-22 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7A3G
 
 | | Crystal structure of DPP8 in complex with a 4-oxo-b-lactam based inhibitor, 91 | | 分子名称: | 1-[3-(7,8-dihydro-5~{H}-[1,3]dioxolo[4,5-g]isoquinolin-6-ylmethyl)phenyl]-3,3-diethyl-azetidine-2,4-dione, CHLORIDE ION, Dipeptidyl peptidase 8, ... | | 著者 | Ross, B.H, Huber, R. | | 登録日 | 2020-08-18 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 2022
|
|
7A3L
 
 | |
7A3J
 
 | |
8API
 
 | | THE S VARIANT OF HUMAN ALPHA1-ANTITRYPSIN, STRUCTURE AND IMPLICATIONS FOR FUNCTION AND METABOLISM | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-1 ANTITRYPSIN (CHAIN A), ... | | 著者 | Loebermann, H, Tokuoka, R, Deisenhofer, J, Huber, R. | | 登録日 | 1988-09-08 | | 公開日 | 1990-10-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The S variant of human alpha 1-antitrypsin, structure and implications for function and metabolism.
Protein Eng., 2, 1989
|
|
7A3F
 
 | | Crystal structure of apo DPP9 | | 分子名称: | Dipeptidyl peptidase 9, GLYCEROL, PHOSPHATE ION | | 著者 | Ross, B.H, Huber, R. | | 登録日 | 2020-08-18 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Discovery and Development of 4-Oxo-beta-Lactams as Novel Inhibitors of Dipeptidyl Peptidases 8 and 9
To Be Published
|
|
7A3I
 
 | |
7A3K
 
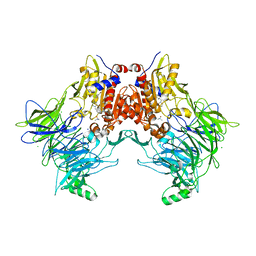 | |
1A23
 
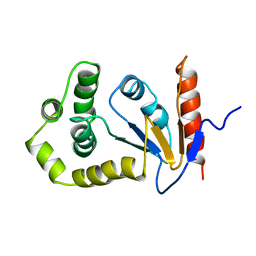 | | SOLUTION NMR STRUCTURE OF REDUCED DSBA FROM ESCHERICHIA COLI, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DSBA | | 著者 | Schirra, H.J, Renner, C, Czisch, M, Huber-Wunderlich, M, Holak, T.A, Glockshuber, R. | | 登録日 | 1998-01-15 | | 公開日 | 1998-09-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of reduced DsbA from Escherichia coli in solution.
Biochemistry, 37, 1998
|
|
1A24
 
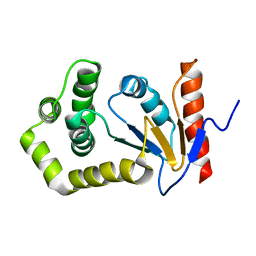 | | SOLUTION NMR STRUCTURE OF REDUCED DSBA FROM ESCHERICHIA COLI, FAMILY OF 20 STRUCTURES | | 分子名称: | DSBA | | 著者 | Schirra, H.J, Renner, C, Czisch, M, Huber-Wunderlich, M, Holak, T.A, Glockshuber, R. | | 登録日 | 1998-01-15 | | 公開日 | 1998-09-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of reduced DsbA from Escherichia coli in solution.
Biochemistry, 37, 1998
|
|
6Z6O
 
 | | HDAC-TC | | 分子名称: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | 著者 | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | 登録日 | 2020-05-28 | | 公開日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
5AZU
 
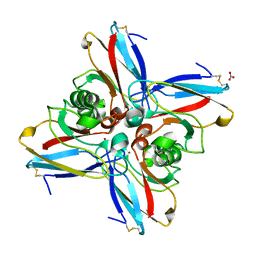 | |
6Z6F
 
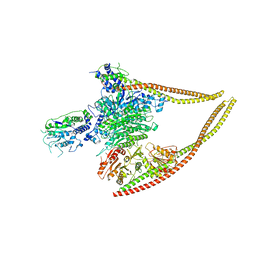 | | HDAC-PC | | 分子名称: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | 著者 | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | 登録日 | 2020-05-28 | | 公開日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6H
 
 | | HDAC-DC | | 分子名称: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | 著者 | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | 登録日 | 2020-05-28 | | 公開日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (8.55 Å) | | 主引用文献 | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6P
 
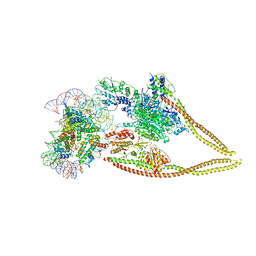 | | HDAC-PC-Nuc | | 分子名称: | DNA (145-MER), HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, ... | | 著者 | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | 登録日 | 2020-05-28 | | 公開日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (4.43 Å) | | 主引用文献 | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
