4UR9
 
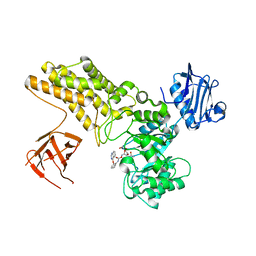 | | Structure of ligand bound glycosylhydrolase | | Descriptor: | 4-ethoxyquinazoline, CALCIUM ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, ... | | Authors: | Darby, J.F, Landstroem, J, Roth, C, He, Y, Schultz, M, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2014-06-27 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Selective Small-Molecule Activators of a Bacterial Glycoside Hydrolase.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6H75
 
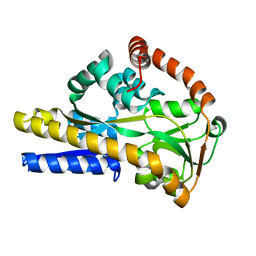 | | SiaP A11N in complex with Neu5Ac (RT) | | Descriptor: | N-acetyl-beta-neuraminic acid, Sialic acid-binding periplasmic protein SiaP | | Authors: | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
6H76
 
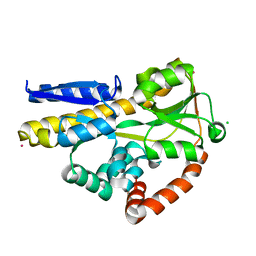 | | SiaP in complex with Neu5Ac (RT) | | Descriptor: | CESIUM ION, CHLORIDE ION, N-acetyl-beta-neuraminic acid, ... | | Authors: | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
1HG7
 
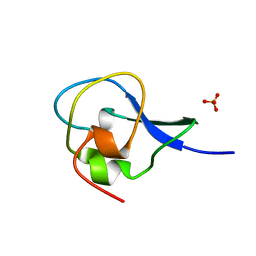 | | High resolution structure of HPLC-12 type III antifreeze protein from Ocean Pout Macrozoarces americanus | | Descriptor: | HPLC-12 TYPE III ANTIFREEZE PROTEIN, SULFATE ION | | Authors: | Antson, A.A, Smith, D.J, Roper, D.I, Lewis, S, Caves, L.S.D, Verma, C.S, Buckley, S.L, Lillford, P.J, Hubbard, R.E. | | Deposit date: | 2000-12-13 | | Release date: | 2001-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Understanding the Mechanism of Ice Binding by Type III Antifreeze Protein
J.Mol.Biol., 305, 2001
|
|
4TLI
 
 | | THERMOLYSIN (25% ISOPROPANOL SOAKED CRYSTALS) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1998-10-29 | | Release date: | 2000-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
8AHH
 
 | |
8AHE
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | SULFATE ION, UDP-N-acetylglucosamine 2-epimerase, ~{N},5-dimethyl-1-phenyl-pyrazole-4-sulfonamide | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8AHG
 
 | |
8AHF
 
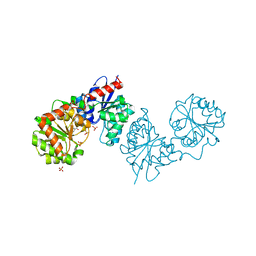 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | (2~{R},4~{S})-4-[bis(fluoranyl)methoxy]-~{N}-methyl-1-(2~{H}-pyrazolo[4,3-b]pyridin-6-ylcarbonyl)pyrrolidine-2-carboxamide, SULFATE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8AHI
 
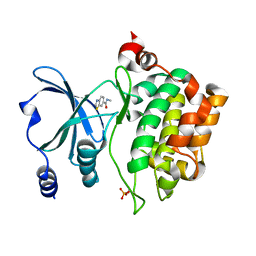 | |
8TLI
 
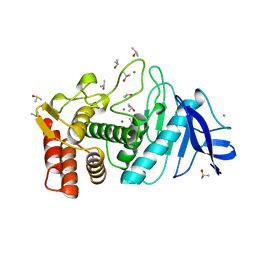 | | THERMOLYSIN (100% ISOPROPANOL SOAKED CRYSTALS) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1998-10-30 | | Release date: | 2000-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
7TLI
 
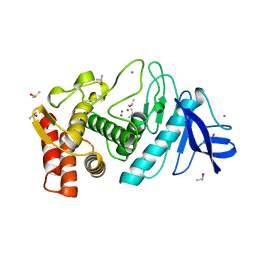 | | THERMOLYSIN (90% ISOPROPANOL SOAKED CRYSTALS) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1998-10-30 | | Release date: | 2000-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
5MI7
 
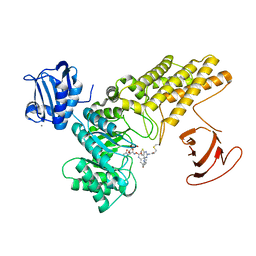 | | BtGH84 mutant with covalent modification by MA4 in complex with PUGNAc | | Descriptor: | CALCIUM ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, O-GlcNAcase BT_4395, ... | | Authors: | Darby, J.F, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2016-11-27 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Increase of enzyme activity through specific covalent modification with fragments.
Chem Sci, 8, 2017
|
|
5MI5
 
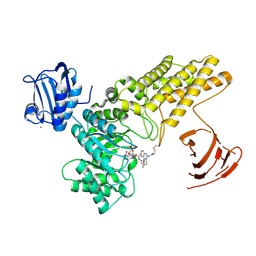 | | BtGH84 mutant with covalent modification by MA3 in complex with PUGNAc | | Descriptor: | CALCIUM ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, O-GlcNAcase BT_4395, ... | | Authors: | Darby, J.F, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2016-11-27 | | Release date: | 2017-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Increase of enzyme activity through specific covalent modification with fragments.
Chem Sci, 8, 2017
|
|
5MI4
 
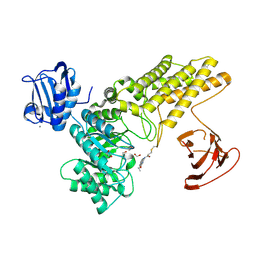 | | BtGH84 mutant with covalent modification by MA3 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, O-GlcNAcase BT_4395, ... | | Authors: | Darby, J.F, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2016-11-27 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Increase of enzyme activity through specific covalent modification with fragments.
Chem Sci, 8, 2017
|
|
5MI6
 
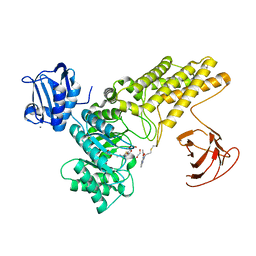 | | BtGH84 mutant with covalent modification by MA3 in complex with Thiamet G | | Descriptor: | (3AR,5R,6S,7R,7AR)-2-(ETHYLAMINO)-5-(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Darby, J.F, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2016-11-27 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Increase of enzyme activity through specific covalent modification with fragments.
Chem Sci, 8, 2017
|
|
3TLI
 
 | | THERMOLYSIN (10% ISOPROPANOL SOAKED CRYSTALS) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1998-10-28 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
5TLI
 
 | | THERMOLYSIN (60% ISOPROPANOL SOAKED CRYSTALS) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1998-10-29 | | Release date: | 2000-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
6QZ7
 
 | | Structure of MBP-Mcl-1 in complex with compound 8b | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZB
 
 | | Structure of Mcl-1 in complex with compound 8d | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(2-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QXJ
 
 | | Structure of MBP-Mcl-1 in complex with compound 6a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-5-yl)thieno[2,3-d]pyrimidin-4-yl]amino]propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-07 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
1BBJ
 
 | |
2BJ4
 
 | | ESTROGEN RECEPTOR ALPHA LBD IN COMPLEX WITH A PHAGE-DISPLAY DERIVED PEPTIDE ANTAGONIST | | Descriptor: | 4-HYDROXYTAMOXIFEN, ESTROGEN RECEPTOR, PEPTIDE ANTAGONIST | | Authors: | Kong, E, Heldring, N, Gustafsson, J.A, Treuter, E, Hubbard, R.E, Pike, A.C.W. | | Deposit date: | 2005-01-28 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Delineation of a Unique Protein-Protein Interaction Site on the Surface of the Estrogen Receptor
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BT0
 
 | | Novel, potent small molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design | | Descriptor: | 4-[4-(2,3-DIHYDRO-1,4-BENZODIOXIN-6-YL)-3-METHYL-1H-PYRAZOL-5-YL]-6-ETHYLBENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP90-ALPHA | | Authors: | Dymock, B.W, Barril, X, Brough, P.A, Cansfield, J.E, Massey, A, McDonald, E, Hubbard, R.E, Surgenor, A, Roughley, S.D, Webb, P, Workman, P, Wright, L, Drysdale, M.J. | | Deposit date: | 2005-05-24 | | Release date: | 2005-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel, potent small-molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design.
J. Med. Chem., 48, 2005
|
|
2BSM
 
 | | Novel, potent small molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design | | Descriptor: | 5-(5-CHLORO-2,4-DIHYDROXYPHENYL)-N-ETHYL-4-(4-METHOXYPHENYL)-1H-PYRAZOLE-3-CARBOXAMIDE, HEAT SHOCK PROTEIN HSP90-ALPHA | | Authors: | Dymock, B.W, Barril, X, Brough, P.A, Cansfield, J.E, Massey, A, McDonald, E, Hubbard, R.E, Surgenor, A, Roughley, S.D, Webb, P, Workman, P, Wright, L, Drysdale, M.J. | | Deposit date: | 2005-05-23 | | Release date: | 2005-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Novel, potent small-molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design.
J. Med. Chem., 48, 2005
|
|
