5Z9Q
 
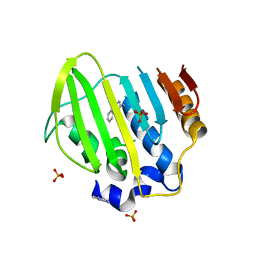 | | Bacterial GyrB ATPase domain in complex with a chemical fragment | | Descriptor: | 1H-benzimidazol-2-amine, 5-phenyl-1H-pyrazol-3-amine, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z9B
 
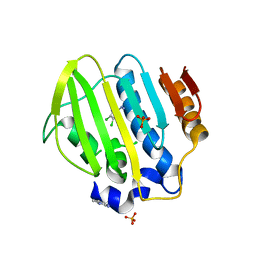 | | Bacterial GyrB ATPase domain in complex with (3,4-dichlorophenyl)hydrazine | | Descriptor: | (3,4-dichlorophenyl)hydrazine, 1H-benzimidazol-2-amine, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-02-02 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z9M
 
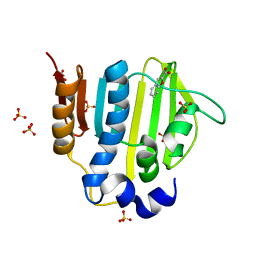 | |
5Z4H
 
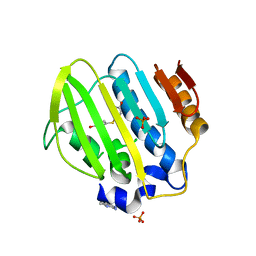 | | Bacterial GyrB ATPase domain in complex with a chemical fragment | | Descriptor: | 1H-benzimidazol-2-amine, 4-(4-hydroxyphenyl)sulfanylphenol, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-01-11 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z9F
 
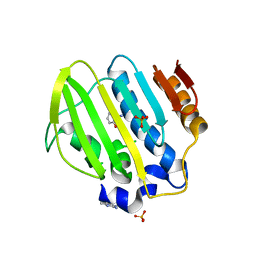 | | Bacterial GyrB ATPase domain in complex with a chemical fragment | | Descriptor: | 1H-benzimidazol-2-amine, 2-hydroxybenzonitrile, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-02-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z9L
 
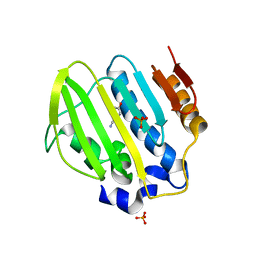 | | Bacterial GyrB ATPase domain in complex with a chemical fragment | | Descriptor: | 1H-benzimidazol-2-amine, 2-fluoro-4-hydroxybenzonitrile, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-02-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z4O
 
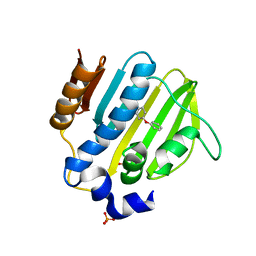 | |
5Z9P
 
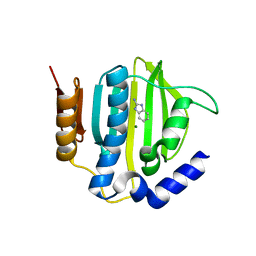 | |
2OFV
 
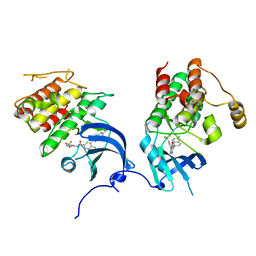 | | crystal structure of aminoquinazoline 1 bound to Lck | | Descriptor: | 3-(2-AMINOQUINAZOLIN-6-YL)-4-METHYL-N-[3-(TRIFLUOROMETHYL)PHENYL]BENZAMIDE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2007-01-04 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Aminoquinazolines as Potent, Orally Bioavailable Inhibitors of Lck: Synthesis, SAR, and in Vivo Anti-Inflammatory Activity
J.Med.Chem., 49, 2006
|
|
2OFU
 
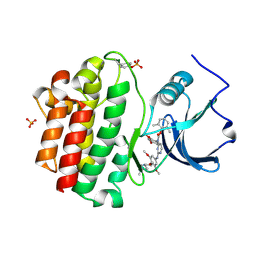 | | x-ray crystal structure of 2-aminopyrimidine carbamate 43 bound to Lck | | Descriptor: | 2,6-DIMETHYLPHENYL 2-(3,5-DIMETHOXY-4-(3-(4-METHYLPIPERAZIN-1-YL)PROPOXY)PHENYLAMINO)PYRIMIDIN- 4-YL(2,4-DIMETHOXYPHENYL)CARBAMATE, Proto-oncogene tyrosine-protein kinase LCK, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2007-01-04 | | Release date: | 2007-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel 2-Aminopyrimidine Carbamates as Potent and Orally Active Inhibitors of Lck: Synthesis, SAR, and in Vivo Antiinflammatory Activity
J.Med.Chem., 49, 2006
|
|
2OG8
 
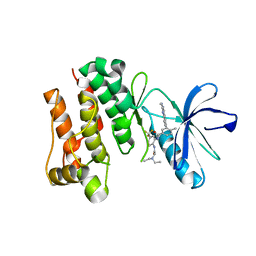 | | crystal structure of aminoquinazoline 36 bound to Lck | | Descriptor: | N-{2-[(N,N-DIETHYLGLYCYL)AMINO]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[2-(METHYLAMINO)QUINAZOLIN-6-YL]BENZAMIDE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Aminoquinazolines as Potent, Orally Bioavailable Inhibitors of Lck: Synthesis, SAR, and in Vivo Anti-Inflammatory Activity
J.Med.Chem., 49, 2006
|
|
2JHB
 
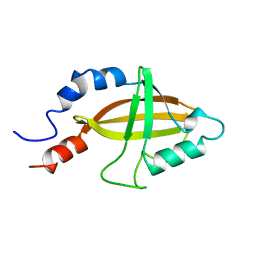 | | CORE BINDING FACTOR BETA | | Descriptor: | PROTEIN (CORE BINDING FACTOR BETA) | | Authors: | Huang, X, Peng, J, Speck, N.A, Bushweller, J.H. | | Deposit date: | 1999-03-17 | | Release date: | 1999-07-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of core binding factor beta and map of the CBF alpha binding site.
Nat.Struct.Biol., 6, 1999
|
|
2R8U
 
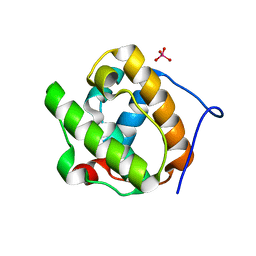 | |
7VOO
 
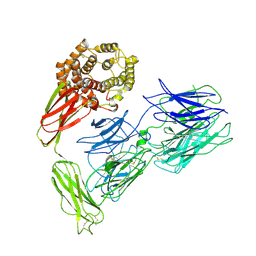 | | Induced alpha-2-macroglobulin monomer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin, ... | | Authors: | Huang, X, Wang, Y, Ping, Z. | | Deposit date: | 2021-10-14 | | Release date: | 2022-10-19 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures reveal the dynamic transformation of human alpha-2-macroglobulin working as a protease inhibitor.
Sci China Life Sci, 65, 2022
|
|
7VON
 
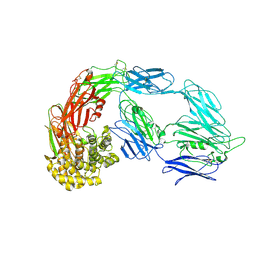 | | Native alpha-2-macroglobulin monomer | | Descriptor: | Alpha-2-macroglobulin | | Authors: | Huang, X, Wang, Y. | | Deposit date: | 2021-10-14 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-EM structures reveal the dynamic transformation of human alpha-2-macroglobulin working as a protease inhibitor.
Sci China Life Sci, 65, 2022
|
|
7VM8
 
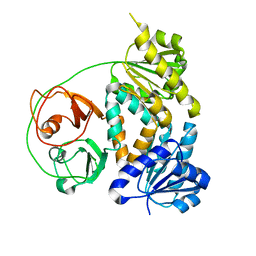 | | Crystal structure of the MtDMI1 gating ring | | Descriptor: | Ion channel DMI1 | | Authors: | Huang, X, Zhang, P. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.034 Å) | | Cite: | Constitutive activation of a nuclear-localized calcium channel complex in Medicago truncatula.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2JXW
 
 | |
4WBE
 
 | |
4WBP
 
 | |
3M4O
 
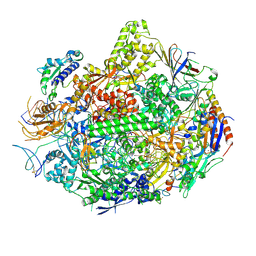 | | RNA polymerase II elongation complex B | | Descriptor: | DNA (28-MER), DNA (5'-D(P*GP*TP*GP*GP*TP*TP*AP*TP*GP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Zhu, G, Huang, X, Lippard, S.J. | | Deposit date: | 2010-03-11 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4H07
 
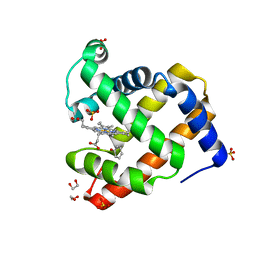 | | Complex of G65T Myoglobin with Phenol in its Proximal Cavity | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, PHENOL, ... | | Authors: | Lebioda, L, Lovelace, L.L, Celeste, L.R, Huang, X, Wang, C, Shengfang, S, Dawson, J.H. | | Deposit date: | 2012-09-07 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Complex of myoglobin with phenol bound in a proximal cavity.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3ZIF
 
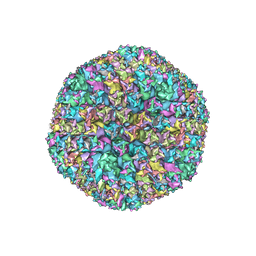 | | Cryo-EM structures of two intermediates provide insight into adenovirus assembly and disassembly | | Descriptor: | HEXON PROTEIN, PENTON PROTEIN, PIX, ... | | Authors: | Cheng, L, Huang, X, Li, X, Xiong, W, Sun, W, Yang, C, Zhang, K, Wang, Y, Liu, H, Ji, G, Sun, F, Zheng, C, Zhu, P. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Em Structures of Two Bovine Adenovirus Type 3 Intermediates
Virology, 450, 2014
|
|
1NOQ
 
 | | e-motif structure | | Descriptor: | 5'-D(*CP*CP*GP*CP*CP*G)-3' | | Authors: | Zheng, M, Huang, X, Smith, G.K, Yang, X, Gao, X. | | Deposit date: | 2003-01-16 | | Release date: | 2003-02-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Genetically unstable CXG repeats are structurally dynamic and have a high propensity for folding.
An NMR and UV spectroscopic study.
J.Mol.Biol., 264, 1996
|
|
1HWV
 
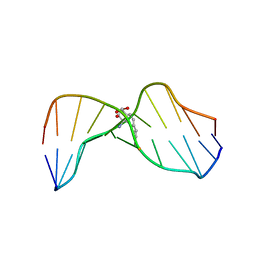 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1S)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-10 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
1HX4
 
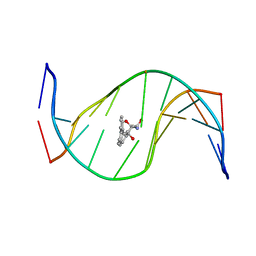 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1R)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-11 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
