6L82
 
 | | Crystal structure of Chaetomium GCP5 N-terminus and Mozart1 | | Descriptor: | Mozart1, Spindle pole body component | | Authors: | Huang, T.L, Wang, H.J, Wang, S.W, Hsia, K.C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.24103618 Å) | | Cite: | Promiscuous Binding of Microprotein Mozart1 to gamma-Tubulin Complex Mediates Specific Subcellular Targeting to Control Microtubule Array Formation.
Cell Rep, 31, 2020
|
|
6M33
 
 | |
4DNS
 
 | | Crystal structure of Bermuda grass isoallergen BG60 provides insight into the various cross-allergenicity of the pollen group 4 allergens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FAD-linked oxidoreductase BG60, ... | | Authors: | Huang, T.H, Peng, H.J, Su, S.N, Liaw, S.H. | | Deposit date: | 2012-02-08 | | Release date: | 2012-12-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Various cross-reactivity of the grass pollen group 4 allergens: crystallographic study of the Bermuda grass isoallergen Cyn d 4.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2MI8
 
 | |
6L81
 
 | | Crystal structure of Homo sapiens GCP5 N-terminus and Mozart1 | | Descriptor: | Gamma-tubulin complex component 5, Mitotic-spindle organizing protein 1 | | Authors: | Huang, T.L, Wang, H.J, Wang, S.W, Hsia, K.C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.19651 Å) | | Cite: | Promiscuous Binding of Microprotein Mozart1 to gamma-Tubulin Complex Mediates Specific Subcellular Targeting to Control Microtubule Array Formation.
Cell Rep, 31, 2020
|
|
6L80
 
 | | Crystal structure of pombe Mod21 N-terminus and Mozart1 | | Descriptor: | Gamma-tubulin complex subunit mod21, Mitotic-spindle organizing protein 1 | | Authors: | Huang, T.L, Wang, H.J, Wang, S.W, Hsia, K.C. | | Deposit date: | 2019-11-03 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.00049758 Å) | | Cite: | Promiscuous Binding of Microprotein Mozart1 to gamma-Tubulin Complex Mediates Specific Subcellular Targeting to Control Microtubule Array Formation.
Cell Rep, 31, 2020
|
|
6L7R
 
 | | Crystal structure of Chaetomium GCP3 N-terminus and Mozart1 | | Descriptor: | Mozart1, Putative spindle pole body component alp6 protein | | Authors: | Huang, T.L, Wang, H.J, Hsia, K.C. | | Deposit date: | 2019-11-02 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8481313 Å) | | Cite: | Promiscuous Binding of Microprotein Mozart1 to gamma-Tubulin Complex Mediates Specific Subcellular Targeting to Control Microtubule Array Formation.
Cell Rep, 31, 2020
|
|
6DXK
 
 | | Glucocorticoid Receptor in complex with Compound 11 | | Descriptor: | (8S,11R,13S,14S,17S)-11-[4-(dimethylamino)phenyl]-17-(3,3-dimethylbut-1-yn-1-yl)-17-hydroxy-13-methyl-1,2,6,7,8,11,12,13,14,15,16,17-dodecahydro-3H-cyclopenta[a]phenanthren-3-one (non-preferred name), Glucocorticoid receptor | | Authors: | Rew, Y, Du, X, Eksterowicz, J, Zhou, H, Jahchan, N, Zhu, L, Yan, X, Kawai, H, McGee, L.R, Medina, J.C, Huang, T, Chen, C, Zavorotinskaya, T, Sutimantanapi, D, Waszczuk, J, Jackson, E, Huang, E, Ye, Q, Fantin, V.R, Daqing, S. | | Deposit date: | 2018-06-29 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of a Potent and Selective Steroidal Glucocorticoid Receptor Antagonist (ORIC-101).
J. Med. Chem., 61, 2018
|
|
2GF5
 
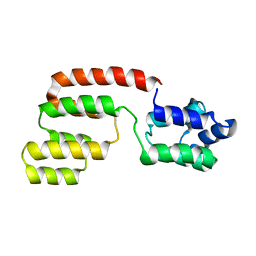 | | Structure of intact FADD (MORT1) | | Descriptor: | FADD protein | | Authors: | Carrington, P.E, Sandu, C, Wei, Y, Hill, J.M, Morisawa, G, Huang, T, Gavathiotis, E, Wei, Y, Werner, M.H. | | Deposit date: | 2006-03-21 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of FADD and Its Mode of Interaction with Procaspase-8
Mol.Cell, 22, 2006
|
|
2GCX
 
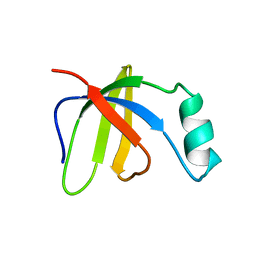 | | Solution Structure of Ferrous Iron Transport Protein A (FeoA) of Klebsiella pneumoniae | | Descriptor: | Ferrous iron transport protein A | | Authors: | Hung, K.-W, Cheng, C.-C, Yu, T.-H, Wang, S.-H, Chang, C.-F, Tsai, S.-F, Huang, T.-H. | | Deposit date: | 2006-03-14 | | Release date: | 2007-03-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ferrous Iron Transport Protein A (FeoA) of Klebsiella pneumoniae
To be Published
|
|
2RR9
 
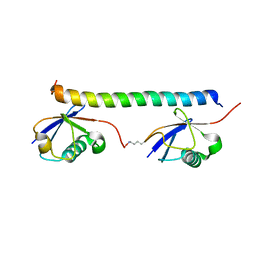 | | The solution structure of the K63-Ub2:tUIMs complex | | Descriptor: | Putative uncharacterized protein UIMC1, ubiquitin | | Authors: | Sekiyama, N, Jee, J, Isogai, S, Akagi, K, Huang, T, Ariyoshi, M, Tochio, H, Shirakawa, M. | | Deposit date: | 2010-06-16 | | Release date: | 2011-07-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the K63-Ub2:tUIMs complex
To be Published
|
|
3WDU
 
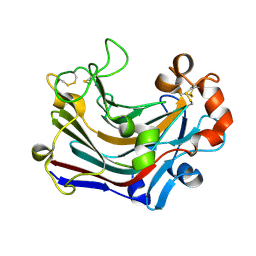 | | The complex structure of PtLic16A with cellobiose | | Descriptor: | Beta-1,3-1,4-glucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
3WDT
 
 | | The apo-form structure of PtLic16A from Paecilomyces thermophila | | Descriptor: | Beta-1,3-1,4-glucanase, SULFATE ION | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
3WH9
 
 | | The ligand-free structure of ManBK from Aspergillus niger BK01 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huang, J.W, Chen, C.C, Huang, C.H, Huang, T.Y, Wu, T.H, Cheng, Y.S, Ko, T.P, Lin, C.Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Analysis and Rational Design to Improve Specific Activity of beta-Mannanase from Aspergillus Niger BK01
To be Published
|
|
3WDY
 
 | | The complex structure of E113A with cellotetraose | | Descriptor: | Beta-1,3-1,4-glucanase, SULFATE ION, beta-D-glucopyranose, ... | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
3WDW
 
 | | The apo-form structure of E113A from Paecilomyces thermophila | | Descriptor: | Beta-1,3-1,4-glucanase, SULFATE ION | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
3WDV
 
 | | The complex structure of PtLic16A with cellotetraose | | Descriptor: | Beta-1,3-1,4-glucanase, SULFATE ION, beta-D-glucopyranose, ... | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
3WDX
 
 | | The complex structure of E113A with glucotriose | | Descriptor: | Beta-1,3-1,4-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
3WP5
 
 | | The crystal structure of mutant CDBFV E109A from Neocallimastix patriciarum | | Descriptor: | CDBFV | | Authors: | Cheng, Y.S, Chen, C.C, Huang, C.H, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-01-09 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 11 xylanase from Neocallimastix patriciarum: insights into the molecular basis of a thermophilic enzyme.
J.Biol.Chem., 289, 2014
|
|
1K8O
 
 | | Solution Structure of the Lipoic Acid-Bearing Domain of the E2 component of Human, Mitochondrial Branched-Chain alpha-Ketoacid Dehydrogenase | | Descriptor: | E2 component of Branched-Chain alpha-Ketoacid Dehydrogenase | | Authors: | Chang, C.-F, Chou, H.-T, Chuang, J.L, Chuang, D.T, Huang, T.-h. | | Deposit date: | 2001-10-24 | | Release date: | 2001-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the lipoic acid-bearing domain of human mitochondrial branched-chain alpha-keto acid dehydrogenase complex
J.Biol.Chem., 277, 2002
|
|
3WP6
 
 | | The complex structure of CDBFV E109A with xylotriose | | Descriptor: | CDBFV, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Cheng, Y.S, Chen, C.C, Huang, C.H, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-01-09 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 11 xylanase from Neocallimastix patriciarum: insights into the molecular basis of a thermophilic enzyme.
J.Biol.Chem., 289, 2014
|
|
3WP4
 
 | | The crystal structure of native CDBFV from Neocallimastix patriciarum | | Descriptor: | CDBFV, SULFATE ION | | Authors: | Cheng, Y.S, Chen, C.C, Huang, C.H, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-01-09 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 11 xylanase from Neocallimastix patriciarum: insights into the molecular basis of a thermophilic enzyme.
J.Biol.Chem., 289, 2014
|
|
1K8M
 
 | | Solution Structure of the Lipoic Acid-Bearing Domain of the E2 component of Human, Mitochondrial Branched-Chain alpha-Ketoacid Dehydrogenase | | Descriptor: | E2 component of Branched-Chain alpha-Ketoacid Dehydrogenase | | Authors: | Chang, C.-F, Chou, H.-T, Chuang, J.L, Chuang, D.T, Huang, T.-h. | | Deposit date: | 2001-10-24 | | Release date: | 2001-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the lipoic acid-bearing domain of human mitochondrial branched-chain alpha-keto acid dehydrogenase complex
J.Biol.Chem., 277, 2002
|
|
1ZWV
 
 | |
2WIB
 
 | | Crystal Structures of the N-terminal Intracellular Domain of FeoB from Klebsiella Pneumoniae in GDP binding state | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Hung, K.-W, Chang, Y.-W, Chen, J.-H, Chen, Y.-C, Sun, Y.-J, Hsiao, C.-D, Huang, T.-H. | | Deposit date: | 2009-05-09 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural Fold, Conservation and Fe(II) Binding of the Intracellular Domain of Prokaryote Feob.
J.Struct.Biol., 170, 2010
|
|
