8GXN
 
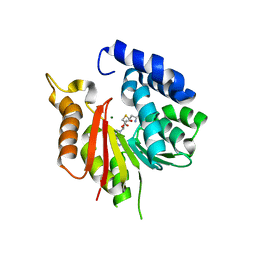 | | The crystal structure of CsFAOMT2 in complex with SAH | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, caffeyl-CoA-O-methyltransferase | | Authors: | Zhang, Z.M, Zhou, Y.E, Huang, H.S. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Characterization of two O-methyltransferases involved in the biosynthesis of O-methylated catechins in tea plant.
Nat Commun, 14, 2023
|
|
1RTD
 
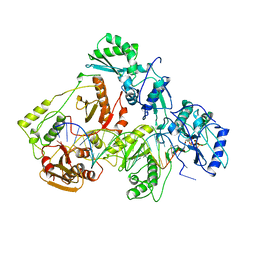 | | STRUCTURE OF A CATALYTIC COMPLEX OF HIV-1 REVERSE TRANSCRIPTASE: IMPLICATIONS FOR NUCLEOSIDE ANALOG DRUG RESISTANCE | | Descriptor: | DNA PRIMER FOR REVERSE TRANSCRIPTASE, DNA TEMPLATE FOR REVERSE TRANSCRIPTASE, MAGNESIUM ION, ... | | Authors: | Chopra, R, Huang, H, Verdine, G.L, Harrison, S.C. | | Deposit date: | 1998-08-26 | | Release date: | 1998-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance.
Science, 282, 1998
|
|
4AZS
 
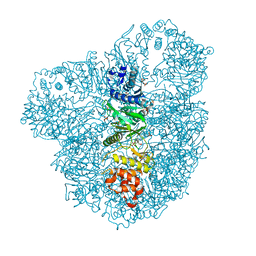 | | High resolution (2.2 A) crystal structure of WbdD. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, METHYLTRANSFERASE WBDD, ... | | Authors: | Hagelueken, G, Huang, H, Naismith, J.H. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Wbdd; a Bifunctional Kinase and Methyltransferase that Regulates the Chain Length of the O Antigen in Escherichia Coli O9A.
Mol.Microbiol., 86, 2012
|
|
4AZW
 
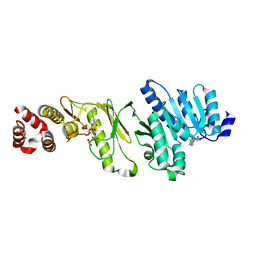 | | Crystal structure of monomeric WbdD. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Hagelueken, G, Huang, H, Naismith, J.H. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure of Wbdd; a Bifunctional Kinase and Methyltransferase that Regulates the Chain Length of the O Antigen in Escherichia Coli O9A.
Mol.Microbiol., 86, 2012
|
|
4A4T
 
 | | UNAC Tetraloops: To What Extent Can They Mimic GNRA Tetraloops | | Descriptor: | 5'-R(*GP*GP*AP*CP*CP*CP*GP*GP*CP*UP*UP*AP*CP*GP *CP*UP*GP*GP*GP*UP*CP*C)-3' | | Authors: | Zhao, Q, Huang, H, Nagaswamy, U, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unac Tetraloops: To What Extent Can They Mimic Gnra Tetraloops
Biopolymers, 97, 2012
|
|
4A4R
 
 | | UNAC Tetraloops: To What Extent Can They Mimic GNRA Tetraloops | | Descriptor: | 5'-R(*GMP*GP*AP*CP*CP*CP*GP*GP*CP*UP*AP*AP*CP*GP *CP*UP*GP*GP*GP*UP*CP*C)-3' | | Authors: | Zhao, Q, Huang, H, Nagaswamy, U, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unac Tetraloops: To What Extent Can They Mimic Gnra Tetraloops
Biopolymers, 97, 2012
|
|
1F3H
 
 | | X-RAY CRYSTAL STRUCTURE OF THE HUMAN ANTI-APOPTOTIC PROTEIN SURVIVIN | | Descriptor: | SULFATE ION, SURVIVIN, ZINC ION | | Authors: | Verdecia, M.A, Huang, H, Dutil, E, Hunter, T, Noel, J.P. | | Deposit date: | 2000-06-03 | | Release date: | 2000-12-06 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement.
Nat.Struct.Biol., 7, 2000
|
|
8JF3
 
 | | C-Src in complex with compound 9 | | Descriptor: | 2-[4-[4-[bis(oxidanylidene)-$l^5-sulfanyl]oxyphenyl]carbonylpiperazin-1-yl]-6-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]-N-prop-2-ynyl-pyrimidine-4-carboxamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Zhang, Z.M, Huang, H.S. | | Deposit date: | 2023-05-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.84647632 Å) | | Cite: | Global Reactivity Profiling of the Catalytic Lysine in Human Kinome for Covalent Inhibitor Development.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
4IIO
 
 | | Crystal Structure of the Second SH3 Domain of ITSN2 Bound with a Synthetic Peptide | | Descriptor: | CHLORIDE ION, Intersectin-2, SULFATE ION, ... | | Authors: | Dong, A, Guan, X, Huang, H, Gu, J, Tempel, W, Sidhu, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-20 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Second SH3 Domain of ITSN2 Bound with a Synthetic Peptide
TO BE PUBLISHED
|
|
7VKA
 
 | | Crystal Structure of GH3.6 in complex with an inhibitor | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Indole-3-acetic acid-amido synthetase GH3.6, ... | | Authors: | Wang, N, Luo, M, Bao, H, Huang, H. | | Deposit date: | 2021-09-29 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Chemical genetic screening identifies nalacin as an inhibitor of GH3 amido synthetase for auxin conjugation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2I7K
 
 | | Solution Structure of the Bromodomain of Human BRD7 Protein | | Descriptor: | Bromodomain-containing protein 7 | | Authors: | Sun, H, Liu, J, Zhang, J, Huang, H, Wu, J, Shi, Y. | | Deposit date: | 2006-08-31 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4
Biochem.Biophys.Res.Commun., 358, 2007
|
|
6B2E
 
 | | Structure of full length human AMPK (a2b2g1) in complex with a small molecule activator SC4. | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Ngoei, K.R.W, Langendorf, C.G, Ling, N.X.Y, Hoque, A, Johnson, S, Camerino, M.C, Walker, S.R, Bozikis, Y.E, Dite, T.A, Ovens, A.J, Smiles, W.J, Jacobs, R, Huang, H, Parker, M.W, Scott, J.W, Rider, M.H, Kemp, B.E, Foitzik, R.C, Baell, J.B, Oakhill, J.S. | | Deposit date: | 2017-09-19 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Determinants for Small-Molecule Activation of Skeletal Muscle AMPK alpha 2 beta 2 gamma 1 by the Glucose Importagog SC4.
Cell Chem Biol, 25, 2018
|
|
6B1U
 
 | | Structure of full-length human AMPK (a2b1g1) in complex with a small molecule activator SC4 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Ngoei, K.R.W, Langendorf, C.G, Ling, N.X.Y, Hoque, A, Johnson, S, Camerino, M.C, Walker, S.R, Bozikis, Y.E, Dite, T.A, Ovens, A.J, Smiles, W.J, Jacobs, R, Huang, H, Parker, M.W, Scott, J.W, Rider, M.H, Kemp, B.E, Foitzik, R.C, Baell, J.B, Oakhill, J.S. | | Deposit date: | 2017-09-19 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural Determinants for Small-Molecule Activation of Skeletal Muscle AMPK alpha 2 beta 2 gamma 1 by the Glucose Importagog SC4.
Cell Chem Biol, 25, 2018
|
|
8GW4
 
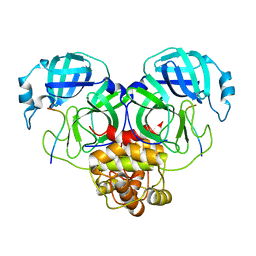 | | SARS-CoV-2 Mpro 1-302/C145A in complex with peptide 8-1 | | Descriptor: | Replicase polyprotein 1ab, peptide 8-1 | | Authors: | Liu, M, Huang, H. | | Deposit date: | 2022-09-16 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The S1'-S3' Pocket of the SARS-CoV-2 Main Protease Is Critical for Substrate Selectivity and Can Be Targeted with Covalent Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8GWS
 
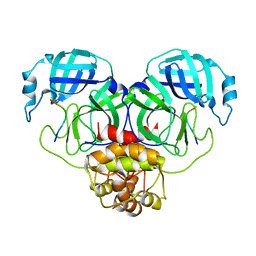 | | SARS-CoV-2 Mpro 1-302 c145a in complex with peptide 4 | | Descriptor: | Replicase polyprotein 1ab, VAL-LYS-LEU-GLN-ALA-ILE-PHE-ARG | | Authors: | Liu, M, Fu, Z, Huang, H. | | Deposit date: | 2022-09-17 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The S1'-S3' Pocket of the SARS-CoV-2 Main Protease Is Critical for Substrate Selectivity and Can Be Targeted with Covalent Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8GXO
 
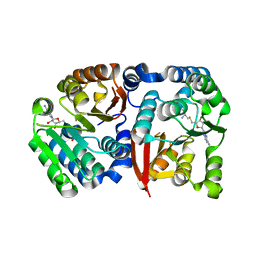 | |
7WFC
 
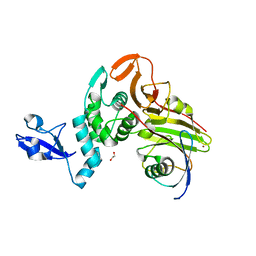 | | X-ray structure of HKU1-PLP2(Cys109Ser) catalytic mutant in complex with free ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, 60S ribosomal protein L40, Papain-like protease, ... | | Authors: | Xiong, Y.X, Fu, Z.Y, Huang, H. | | Deposit date: | 2021-12-26 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The substrate selectivity of papain-like proteases from human-infecting coronaviruses correlates with innate immune suppression.
Sci.Signal., 16, 2023
|
|
4IIM
 
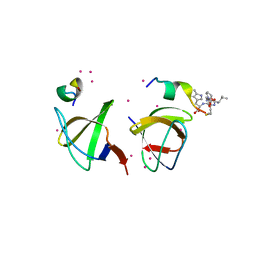 | | Crystal structure of the Second SH3 Domain of ITSN1 bound with a synthetic peptide | | Descriptor: | Intersectin-1, UNKNOWN ATOM OR ION, peptide ligand | | Authors: | Dong, A, Guan, X, Huang, H, Wernimont, A, Gu, J, Sidhu, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Second SH3 Domain of ITSN1 bound with a synthetic peptide
To be Published
|
|
6LOJ
 
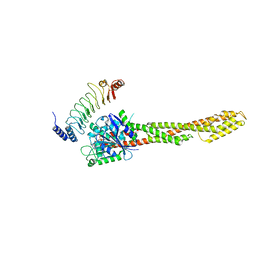 | | The complex structure of IpaH9.8-LRR and hGBP1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanylate-binding protein 1, Invasion plasmid antigen | | Authors: | Ye, Y, Huang, H. | | Deposit date: | 2020-01-06 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.72 Å) | | Cite: | Substrate-binding destabilizes the hydrophobic cluster to relieve the autoinhibition of bacterial ubiquitin ligase IpaH9.8.
Commun Biol, 3, 2020
|
|
3PJZ
 
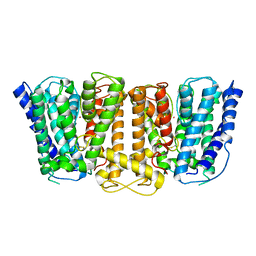 | | Crystal Structure of the Potassium Transporter TrkH from Vibrio parahaemolyticus | | Descriptor: | POTASSIUM ION, Potassium uptake protein TrkH | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2010-11-10 | | Release date: | 2011-01-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Crystal structure of a potassium ion transporter, TrkH.
Nature, 471, 2011
|
|
6LOL
 
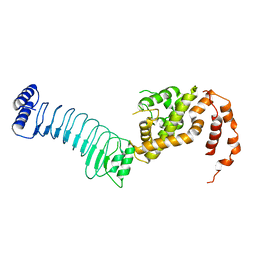 | | The crystal structure of full length IpaH9.8 | | Descriptor: | E3 ubiquitin-protein ligase ipaH9.8 | | Authors: | Ye, Y, Huang, H. | | Deposit date: | 2020-01-06 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Substrate-binding destabilizes the hydrophobic cluster to relieve the autoinhibition of bacterial ubiquitin ligase IpaH9.8.
Commun Biol, 3, 2020
|
|
6J7B
 
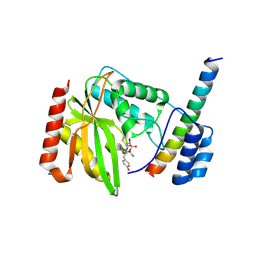 | | Crystal structure of VASH1-SVBP in complex with epoY | | Descriptor: | N-[(3R)-4-ethoxy-3-hydroxy-4-oxobutanoyl]-L-tyrosine, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Wang, N, Bao, H, Huang, H, Wu, B. | | Deposit date: | 2019-01-17 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.618 Å) | | Cite: | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
3QYP
 
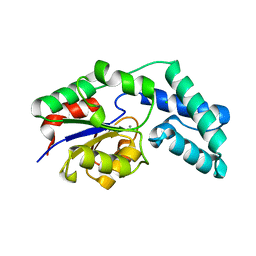 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, glu47asn mutant complexed with calcium and phosphate | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-03-03 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QUC
 
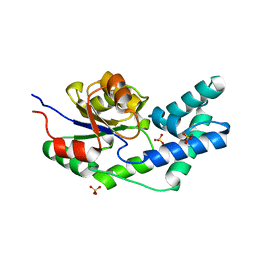 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, glu47asn mutant complexed with sulfate | | Descriptor: | INORGANIC PYROPHOSPHATASE, SULFATE ION | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
5H2T
 
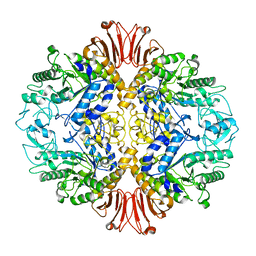 | |
