2XWT
 
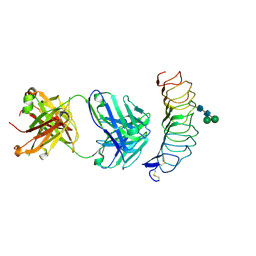 | | CRYSTAL STRUCTURE OF THE TSH RECEPTOR IN COMPLEX WITH A BLOCKING TYPE TSHR AUTOANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, THYROID BLOCKING HUMAN AUTOANTIBODY K1-70 HEAVY CHAIN, ... | | Authors: | Sanders, J, Sanders, P, Young, S, Kabelis, K, Baker, S, Sullivan, A, Evans, M, Clark, J, Wilmot, J, Hu, X, Roberts, E, Powell, M, Nunez Miguel, R, Furmaniak, J, Rees Smith, B. | | Deposit date: | 2010-11-05 | | Release date: | 2011-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Tsh Receptor (Tshr) Bound to a Blocking-Type Tshr Autoantibody.
J.Mol.Endocrinol., 46, 2011
|
|
6S8Y
 
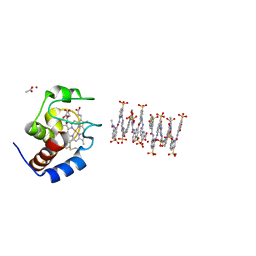 | | Crystal structure of cytochrome c in complex with a sulfonated quinoline-derived foldamer | | Descriptor: | 8-acetamido-2-[[2-[[2-[[2-[[2-[[2-[[2-[(2-carboxy-4-sulfonato-quinolin-8-yl)carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]quinoline-4-sulfonate, ACETATE ION, Cytochrome c iso-1, ... | | Authors: | Alex, J.M, Corvaglia, V, Hu, X, Engilberge, S, Huc, I, Crowley, P.B. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a protein-aromatic foldamer composite: macromolecular chiral resolution.
Chem.Commun.(Camb.), 55, 2019
|
|
4U7O
 
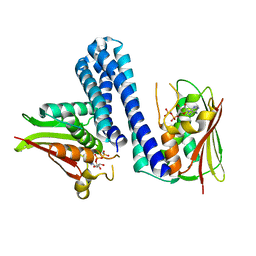 | | Active histidine kinase bound with ATP | | Descriptor: | AMP PHOSPHORAMIDATE, Histidine protein kinase sensor protein | | Authors: | Cai, Y, Hu, X, Sang, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Conformational dynamics of the essential sensor histidine kinase WalK.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4U7N
 
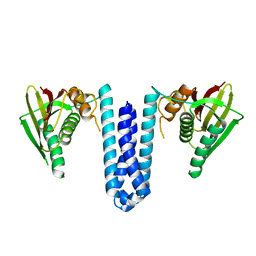 | | Inactive structure of histidine kinase | | Descriptor: | Histidine protein kinase sensor protein | | Authors: | Cai, Y, Hu, X, Sang, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational dynamics of the essential sensor histidine kinase WalK.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1X9Z
 
 | | Crystal structure of the MutL C-terminal domain | | Descriptor: | CHLORIDE ION, DNA mismatch repair protein mutL, GLYCEROL, ... | | Authors: | Guarne, A, Ramon-Maiques, S, Wolff, E.M, Ghirlando, R, Hu, X, Miller, J.H, Yang, W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair
Embo J., 23, 2004
|
|
1YCL
 
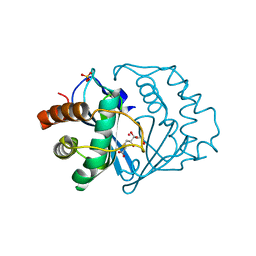 | | Crystal Structure of B. subtilis LuxS in Complex with a Catalytic 2-Ketone Intermediate | | Descriptor: | (S)-2-AMINO-4-[(2S,3R)-2,3,5-TRIHYDROXY-4-OXO-PENTYL]MERCAPTO-BUTYRIC ACID, COBALT (II) ION, S-ribosylhomocysteinase, ... | | Authors: | Rajan, R, Zhu, J, Hu, X, Pei, D, Bell, C.E. | | Deposit date: | 2004-12-22 | | Release date: | 2005-03-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of S-Ribosylhomocysteinase (LuxS) in Complex with a Catalytic 2-Ketone Intermediate.
Biochemistry, 44, 2005
|
|
5O4N
 
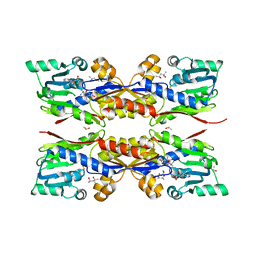 | | Apo HcgC from Methanococcus maripaludis soaked with SAH and pyridinol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-carboxy methyl-4-hydroxy-2-pyridinol, DIMETHYL SULFOXIDE, ... | | Authors: | Wagner, T, Bai, L, Xu, T, Hu, X, Ermler, U, Shima, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Water-Bridged H-Bonding Network Contributes to the Catalysis of the SAM-Dependent C-Methyltransferase HcgC.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5O4J
 
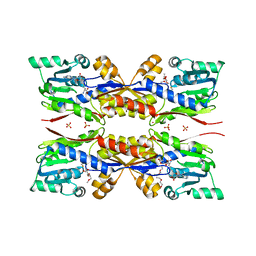 | | HcgC from Methanococcus maripaludis cocrystallized with SAH and pyridinol | | Descriptor: | (3~{E})-3-[(~{E})-3-oxidanylprop-2-enoyl]iminopropanoic acid, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 6-carboxy methyl-4-hydroxy-2-pyridinol, ... | | Authors: | Wagner, T, Bai, L, Xu, T, Hu, X, Ermler, U, Shima, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Water-Bridged H-Bonding Network Contributes to the Catalysis of the SAM-Dependent C-Methyltransferase HcgC.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5O4M
 
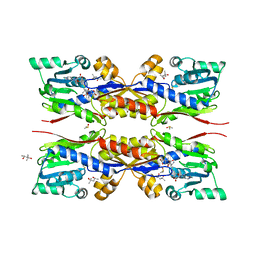 | | Fresh crystals of HcgC from Methanococcus maripaludis cocrystallized with SAH and pyridinol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-carboxy methyl-4-hydroxy-2-pyridinol, DIMETHYL SULFOXIDE, ... | | Authors: | Wagner, T, Bai, L, Xu, T, Hu, X, Ermler, U, Shima, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Water-Bridged H-Bonding Network Contributes to the Catalysis of the SAM-Dependent C-Methyltransferase HcgC.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5O4H
 
 | | HcgC from Methanococcus maripaludis cocrystallized with SAM and pyridinol | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ACETATE ION, HcgC, ... | | Authors: | Wagner, T, Bai, L, Xu, T, Hu, X, Ermler, U, Shima, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Water-Bridged H-Bonding Network Contributes to the Catalysis of the SAM-Dependent C-Methyltransferase HcgC.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4ZFC
 
 | | Crystal structure of AKR1C3 complexed with glicazide | | Descriptor: | Aldo-keto reductase family 1 member C3, N-[(3aR,6aS)-hexahydrocyclopenta[c]pyrrol-2(1H)-ylcarbamoyl]-4-methylbenzenesulfonamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zhrng, X, Hu, X. | | Deposit date: | 2015-04-21 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4YVP
 
 | | Crystal Structure of AKR1C1 complexed with glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4YVX
 
 | | Crystal structure of AKR1C3 complexed with glimepiride | | Descriptor: | 3-ethyl-4-methyl-N-[2-(4-{[(cis-4-methylcyclohexyl)carbamoyl]sulfamoyl}phenyl)ethyl]-2-oxo-2,5-dihydro-1H-pyrrole-1-car boxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4YVV
 
 | | Crystal structure of AKR1C3 complexed with glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
3V36
 
 | | Aldose reductase complexed with glceraldehyde | | Descriptor: | Aldose reductase, D-Glyceraldehyde, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zheng, X, Zhang, L, Chen, Y, Luo, H, Hu, X. | | Deposit date: | 2011-12-13 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Partial inhibition of aldose reductase by nitazoxanide and its molecular basis.
Chemmedchem, 7, 2012
|
|
3V35
 
 | | Aldose reductase complexed with a nitro compound | | Descriptor: | 2-[(5-nitro-1,3-thiazol-2-yl)carbamoyl]phenyl acetate, Aldose reductase, DIMETHYLFORMAMIDE, ... | | Authors: | Zheng, X, Zhang, L, Chen, Y, Luo, H, Hu, X. | | Deposit date: | 2011-12-13 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Partial inhibition of aldose reductase by nitazoxanide and its molecular basis.
Chemmedchem, 7, 2012
|
|
4GQG
 
 | | Crystal structure of AKR1B10 complexed with NADP+ | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Zheng, X, Chen, S, Zhai, J, Hu, X. | | Deposit date: | 2012-08-23 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
3RX3
 
 | | Crystal Structure of Human Aldose Reductase Complexed with Sulindac | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [(1Z)-5-fluoro-2-methyl-1-{4-[methylsulfinyl]benzylidene}-1H-inden-3-yl]acetic acid | | Authors: | Zheng, X, Chen, J, Luo, H, Hu, X. | | Deposit date: | 2011-05-10 | | Release date: | 2011-11-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The molecular basis for inhibition of sulindac and its metabolites towards human aldose reductase.
Febs Lett., 586, 2012
|
|
3RX4
 
 | | Crystal Structure of Human Aldose Reductase complexed with Sulindac Sulfide | | Descriptor: | 2-[(3Z)-6-fluoranyl-2-methyl-3-[(4-methylsulfanylphenyl)methylidene]inden-1-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Chen, J, Luo, H, Hu, X. | | Deposit date: | 2011-05-10 | | Release date: | 2011-11-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The molecular basis for inhibition of sulindac and its metabolites towards human aldose reductase.
Febs Lett., 586, 2012
|
|
3RX2
 
 | | Crystal Structure of Human Aldose Reductase Complexed with Sulindac Sulfone | | Descriptor: | 2-[(3Z)-6-fluoranyl-2-methyl-3-[(4-methylsulfonylphenyl)methylidene]inden-1-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Chen, J, Luo, H, Hu, X. | | Deposit date: | 2011-05-10 | | Release date: | 2011-11-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The molecular basis for inhibition of sulindac and its metabolites towards human aldose reductase
Febs Lett., 586, 2012
|
|
2B3L
 
 | | Crystal structure of type I human methionine aminopeptidase in the apo form | | Descriptor: | ACETIC ACID, GLYCEROL, Methionine aminopeptidase 1, ... | | Authors: | Addlagatta, A, Hu, X, Liu, J.O, Matthews, B.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Functional Differences between Type I and Type II Human Methionine Aminopeptidases(,).
Biochemistry, 44, 2005
|
|
2B3K
 
 | | Crystal structure of Human Methionine Aminopeptidase Type I in the holo form | | Descriptor: | ACETATE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Addlagatta, A, Hu, X, Liu, J.O, Matthews, B.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for the Functional Differences between Type I and Type II Human Methionine Aminopeptidases(,).
Biochemistry, 44, 2005
|
|
2B3H
 
 | | Crystal structure of Human Methionine Aminopeptidase Type I with a third cobalt in the active site | | Descriptor: | CHLORIDE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Addlagatta, A, Hu, X, Liu, J.O, Matthews, B.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Basis for the Functional Differences between Type I and Type II Human Methionine Aminopeptidases(,).
Biochemistry, 44, 2005
|
|
3S3G
 
 | | Crystal Structure of Human Aldose Reductase Complexed with Tolmetin | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Tolmetin | | Authors: | Zheng, X, Chen, J, Luo, H, Hu, X. | | Deposit date: | 2011-05-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis for inhibition of sulindac and its metabolites towards human aldose reductase.
Febs Lett., 586, 2012
|
|
4X2A
 
 | | Crystal structure of mouse glyoxalase I complexed with baicalein | | Descriptor: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhang, H, Zhai, J, Zhang, L, Li, C, Zhao, Y, Hu, X. | | Deposit date: | 2014-11-26 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In Vitro Inhibition of Glyoxalase І by Flavonoids: New Insights from Crystallographic Analysis.
Curr Top Med Chem, 16, 2016
|
|
