8GXN
 
 | |
8GXO
 
 | |
7B76
 
 | | Crystal structure of the effector AvrLm5-9 from Leptosphaeria maculans | | 分子名称: | Avirulence protein LmJ1, IODIDE ION | | 著者 | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | 登録日 | 2020-12-09 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
8HFA
 
 | |
7W0V
 
 | | C4'-SCF3-DT modifeid DNA-DNA duplex | | 分子名称: | DNA (5'-D(*CP*CP*AP*TP*(DSW)P*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | 著者 | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | 登録日 | 2021-11-18 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5JBM
 
 | |
3BV9
 
 | | Structure of Thrombin Bound to the Inhibitor FM19 | | 分子名称: | FM19 inhibitor, GLYCEROL, IODIDE ION, ... | | 著者 | Nieman, M.T, Burke, F, Warnock, M, Zhou, Y, Sweigert, J, Chen, A, Ricketts, D, Lucchesi, B.R, Chen, Z, Di Cera, E, Hilfinger, J, Mosberg, H.I, Schmaier, A.H. | | 登録日 | 2008-01-05 | | 公開日 | 2008-03-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Thrombostatin FM compounds: direct thrombin inhibitors - mechanism of action in vitro and in vivo.
J.Thromb.Haemost., 6, 2008
|
|
6XVD
 
 | | Crystal structure of complex of urokinase and a upain-1 variant(W3F) in pH7.4 condition | | 分子名称: | Urokinase-type plasminogen activator, upain-1-W3F | | 著者 | Xue, G.P, Xie, X, Zhou, Y, Yuan, C, Huang, M.D, Jiang, L.G. | | 登録日 | 2020-01-21 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Insight to the residue in P2 position prevents the peptide inhibitor from being hydrolyzed by serine proteases.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
5KLV
 
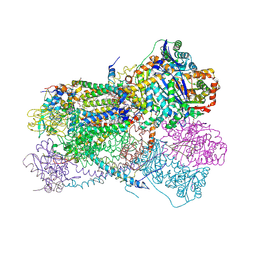 | | Structure of bos taurus cytochrome bc1 with fenamidone inhibited | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, (5S)-5-methyl-2-(methylsulfanyl)-5-phenyl-3-(phenylamino)-3,5-dihydro-4H-imidazol-4-one, 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ... | | 著者 | Xia, D, Esser, L, Zhou, F, Zhou, Y, Xiao, Y, Tang, W.K, Yu, C.A, Qin, Z. | | 登録日 | 2016-06-25 | | 公開日 | 2016-10-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.652 Å) | | 主引用文献 | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
7VUG
 
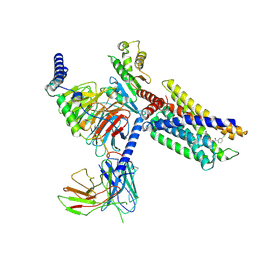 | | Cryo-EM structure of a class A orphan GPCR in complex with Gi | | 分子名称: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Chimera of Endo-1,4-beta-xylanase and Probable G-protein coupled receptor 139, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | 登録日 | 2021-11-02 | | 公開日 | 2021-12-29 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUH
 
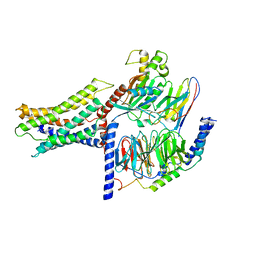 | | Cryo-EM structure of a class A orphan GPCR | | 分子名称: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | 登録日 | 2021-11-02 | | 公開日 | 2021-12-29 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUJ
 
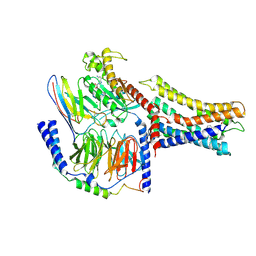 | | Cryo-EM structure of a class A orphan GPCR | | 分子名称: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | 登録日 | 2021-11-02 | | 公開日 | 2021-12-29 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUI
 
 | | Cryo-EM structure of a class A orphan GPCR | | 分子名称: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Chimera of Endo-1,4-beta-xylanase and Probable G-protein coupled receptor 139, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | 登録日 | 2021-11-02 | | 公開日 | 2021-12-29 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
1C1J
 
 | | STRUCTURE OF CADMIUM-SUBSTITUTED PHOSPHOLIPASE A2 FROM AGKISTRONDON HALYS PALLAS AT 2.8 ANGSTROMS RESOLUTION | | 分子名称: | BASIC PHOSPHOLIPASE A2, CADMIUM ION, octyl beta-D-glucopyranoside | | 著者 | Zhang, H.-l, Zhang, Y.-q, Song, S.-y, Zhou, Y, Lin, Z.-j. | | 登録日 | 1999-07-22 | | 公開日 | 2002-07-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of Cadmium-substituted Phospholipase A2 from Agkistrodon halys
Pallas at 2.8 Angstroms Resolution
Protein Pept.Lett., 6, 1999
|
|
8HE4
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | 分子名称: | Chitin deacetylase, ZINC ION, ~{N}-oxidanylnaphthalene-1-carboxamide | | 著者 | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | 登録日 | 2022-11-07 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HE2
 
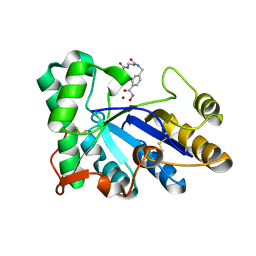 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | 分子名称: | Chitin deacetylase, ZINC ION, tert-butyl N-[3-[[4-(oxidanylcarbamoyl)phenyl]methylamino]-3-oxidanylidene-propyl]carbamate | | 著者 | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | 登録日 | 2022-11-07 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HE1
 
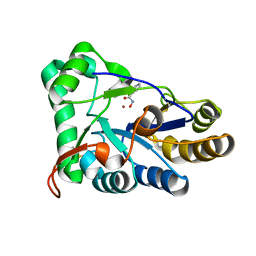 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | 分子名称: | BENZHYDROXAMIC ACID, Chitin deacetylase, ZINC ION | | 著者 | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | 登録日 | 2022-11-07 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HF9
 
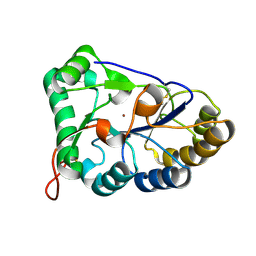 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | 分子名称: | Chitin deacetylase, ZINC ION | | 著者 | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | 登録日 | 2022-11-10 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
6IGY
 
 | |
3FYS
 
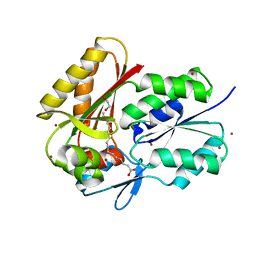 | | Crystal Structure of DegV, a fatty acid binding protein from Bacillus subtilis | | 分子名称: | 1,2-ETHANEDIOL, BROMIDE ION, PALMITIC ACID, ... | | 著者 | Nan, J, Zhou, Y.F, Yang, C. | | 登録日 | 2009-01-23 | | 公開日 | 2009-05-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of a fatty acid-binding protein from Bacillus subtilis determined by sulfur-SAD phasing using in-house chromium radiation
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6JAW
 
 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with a napthalimide derivative | | 分子名称: | 2-[3-(morpholin-4-yl)propyl]-1H-benzo[de]isoquinoline-1,3(2H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, Group II chitinase | | 著者 | Chen, W, Zhou, Y, Yang, Q. | | 登録日 | 2019-01-25 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.981 Å) | | 主引用文献 | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
8GKV
 
 | | Crystal structure of anti-adaptor IraP that regulates RpoS proteolysis | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Shaw, G.X, Gan, J, Suburaman, P, Battesti, A, Zhou, Y.N, Wickner, S, Gottesman, S, Ji, X. | | 登録日 | 2023-03-20 | | 公開日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.351 Å) | | 主引用文献 | Structural and functional study of anti-adaptor IraP-mediated regulation of RpoS proteolysis
to be published
|
|
6JAV
 
 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with a piperidine-thienopyridine derivative | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-chlorophenyl)methyl]sulfanyl}-7-methyl-N-(prop-2-en-1-yl)-7,8-dihydropyrido[4',3':4,5]thieno[2,3-d]pyrimidin-4-amine, Group II chitinase | | 著者 | Chen, W, Zhou, Y, Yang, Q. | | 登録日 | 2019-01-25 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.437 Å) | | 主引用文献 | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
6JAY
 
 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with a dipyrido-pyrimidine derivative | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-1-[(furan-2-yl)methyl]-5-oxo-3-({[(2S)-oxolan-2-yl]methyl}carbamoyl)-5H-dipyrido[1,2-a:2',3'-d]pyrimidin-1-ium, Group II chitinase | | 著者 | Chen, W, Zhou, Y, Yang, Q. | | 登録日 | 2019-01-25 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.498 Å) | | 主引用文献 | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
2AY0
 
 | | Structure of the Lys9Met mutant of the E. coli Proline Utilization A (PutA) DNA-binding domain. | | 分子名称: | Bifunctional putA protein, CHLORIDE ION | | 著者 | Larson, J.D, Schuermann, J.P, Zhou, Y, Jenkins, J.L, Becker, D.F, Tanner, J.J. | | 登録日 | 2005-09-06 | | 公開日 | 2006-08-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of the DNA-binding domain of Escherichia coli proline utilization A flavoprotein and analysis of the role of Lys9 in DNA recognition.
Protein Sci., 15, 2006
|
|
