7X0B
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | 4Fe-4S cluster-binding domain-containing protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02027535 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7WZX
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | (2~{S},4~{S},6~{R})-2-[(2~{S},3~{R},5~{S},6~{R})-3,5-bis(methylamino)-2,4,6-tris(oxidanyl)cyclohexyl]oxy-6-methyl-4-oxidanyl-oxan-3-one, 4Fe-4S cluster-binding domain-containing protein, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2022-02-19 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.980013 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7VOC
 
 | | The crystal structure of a Radical SAM Enzyme BlsE involved in the Biosynthesis of Blasticidin S | | Descriptor: | (2~{S},3~{S},4~{S},5~{R},6~{R})-6-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4,5-tris(oxidanyl)oxane-2-carboxylic acid, Cytosylglucuronate decarboxylase, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2021-10-13 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.62005424 Å) | | Cite: | Radical S -Adenosyl Methionine Enzyme BlsE Catalyzes a Radical-Mediated 1,2-Diol Dehydration during the Biosynthesis of Blasticidin S.
J.Am.Chem.Soc., 144, 2022
|
|
7VOB
 
 | |
5DJ5
 
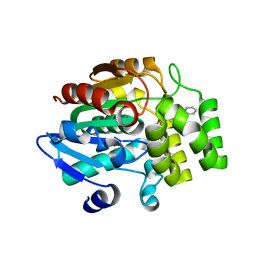 | | Crystal structure of rice DWARF14 in complex with synthetic strigolactone GR24 | | Descriptor: | (3E,3aR,8bS)-3-({[(2R)-4-methyl-5-oxo-2,5-dihydrofuran-2-yl]oxy}methylidene)-3,3a,4,8b-tetrahydro-2H-indeno[1,2-b]furan-2-one, Probable strigolactone esterase D14 | | Authors: | Zhou, X.E, Zhao, L.-H, Yi, W, Wu, Z.-S, Liu, Y, Kang, Y, Hou, L, de Waal, P.W, Li, S, Jiang, Y, Melcher, K, Xu, H.E. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3.
Cell Res., 25, 2015
|
|
4IH1
 
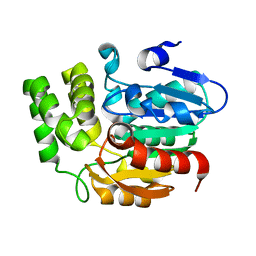 | | Crystal structure of Karrikin Insensitive 2 (KAI2) from Arabidopsis thaliana | | Descriptor: | Hydrolase, alpha/beta fold family protein | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4IH9
 
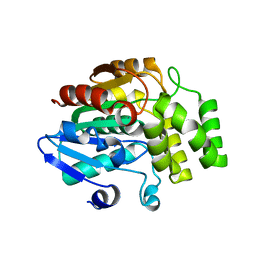 | | Crystal structure of rice DWARF14 (D14) | | Descriptor: | Dwarf 88 esterase | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4IHA
 
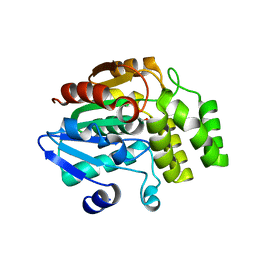 | | Crystal structure of rice DWARF14 (D14) in complex with a GR24 hydrolysis intermediate | | Descriptor: | (2R,3R)-2,4,4-trihydroxy-3-methylbutanal, Dwarf 88 esterase | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4IH4
 
 | | Crystal structure of Arabidopsis DWARF14 orthologue, AtD14 | | Descriptor: | AT3g03990/T11I18_10 | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
8HZV
 
 | | The crystal structure of a Radical SAM Enzyme DesII | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, METHIONINE, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2023-01-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.33001685 Å) | | Cite: | Mechanistic Insights from the Crystal Structure and Computational Analysis of the Radical SAM Deaminase DesII.
Adv Sci, 11, 2024
|
|
8HZY
 
 | | The crystal structure of a Radical SAM Enzyme DesII | | Descriptor: | DesII, IRON/SULFUR CLUSTER, METHIONINE | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2023-01-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.039788 Å) | | Cite: | Mechanistic Insights from the Crystal Structure and Computational Analysis of the Radical SAM Deaminase DesII.
Adv Sci, 11, 2024
|
|
7Y3X
 
 | | Crystal structure of BTG13 mutant (H58F) | | Descriptor: | FE (III) ION, GLYCEROL, Questin oxidase | | Authors: | Hou, X.D, Rao, Y.J. | | Deposit date: | 2022-06-13 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of the Biosynthetic Pathway of Beticolin 1 Reveals a Novel Non-Heme Iron-Dependent Oxygenase for Anthraquinone Ring Cleavage.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Y3Y
 
 | | Crystal structure of BTG13 mutant (T299V) | | Descriptor: | FE (III) ION, GLYCEROL, questin oxidase BTG13 | | Authors: | Hou, X.D, Fu, K, Rao, Y.J. | | Deposit date: | 2022-06-13 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of the Biosynthetic Pathway of Beticolin 1 Reveals a Novel Non-Heme Iron-Dependent Oxygenase for Anthraquinone Ring Cleavage.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Y3W
 
 | |
7YB1
 
 | | Crystal Structure of anthrol reductase (CbAR) in complex with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Versicolorin reductase | | Authors: | Hou, X.D, Rao, Y.J. | | Deposit date: | 2022-06-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural analysis of an anthrol reductase inspires enantioselective synthesis of enantiopure hydroxycycloketones and beta-halohydrins.
Nat Commun, 14, 2023
|
|
7YB2
 
 | | Crystal Structure of anthrol reductase (CbAR) in complex with NADP+ and emodin | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hou, X.D, Rao, Y.J. | | Deposit date: | 2022-06-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of an anthrol reductase inspires enantioselective synthesis of enantiopure hydroxycycloketones and beta-halohydrins.
Nat Commun, 14, 2023
|
|
2R13
 
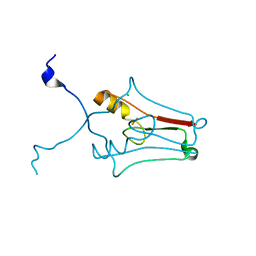 | | Crystal structure of human mitoNEET reveals a novel [2Fe-2S] cluster coordination | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Zinc finger CDGSH domain-containing protein 1 | | Authors: | Hou, X, Liu, R, Ross, S, Smart, E.J, Zhu, H, Gong, W. | | Deposit date: | 2007-08-22 | | Release date: | 2007-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies of human MitoNEET
J.Biol.Chem., 282, 2007
|
|
7VM0
 
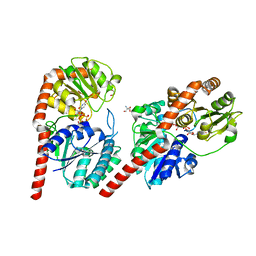 | | Crystal structure of YojK from B.subtilis in complex with UDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Glycosyl transferase family 1, ... | | Authors: | Hou, X.D, Guo, B.D, Rao, Y.J. | | Deposit date: | 2021-10-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly efficient production of rebaudioside D enabled by structure-guided engineering of bacterial glycosyltransferase YojK.
Front Bioeng Biotechnol, 10, 2022
|
|
7EUT
 
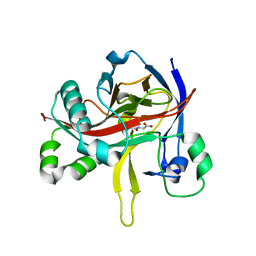 | | Crystal structures of 2-oxoglutarate dependent dioxygenase (CTB9) in complex with N-oxalylglycine | | Descriptor: | 1,2-ETHANEDIOL, 2-oxoglutarate (2-OG)-dependent dioxygenase, COPPER (II) ION, ... | | Authors: | Hou, X.D, Liu, X.Z, Yuan, Z.B, Yin, D.J, Rao, Y.J. | | Deposit date: | 2021-05-18 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Molecular Basis of the Unusual Seven-Membered Methylenedioxy Bridge Formation Catalyzed by Fe(II)/alpha-KG-Dependent Oxygenase CTB9
Acs Catalysis, 12, 2022
|
|
7EUS
 
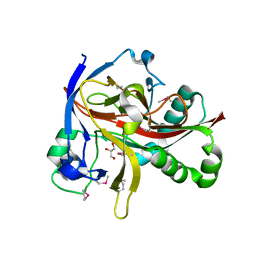 | | Crystal structures of 2-oxoglutarate dependent dioxygenase (CTB9) from Cercospora sp. JNU001 | | Descriptor: | 2-oxoglutarate (2-OG)-dependent dioxygenase, COPPER (II) ION, GLYCEROL | | Authors: | Hou, X.D, Liu, X.Z, Yuan, Z.B, Rao, Y.J. | | Deposit date: | 2021-05-18 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis of the Unusual Seven-Membered Methylenedioxy Bridge Formation Catalyzed by Fe(II)/alpha-KG-Dependent Oxygenase CTB9
Acs Catalysis, 12, 2022
|
|
7EUU
 
 | | Crystal structures of 2-oxoglutarate dependent dioxygenase (CTB9) in complex with N-oxalylglycine and pre-cercosporin | | Descriptor: | 1,2-ETHANEDIOL, 2,6,11-trimethoxy-4,7,9-tris(oxidanyl)-1,12-bis[(2R)-2-oxidanylpropyl]perylene-3,10-dione, 2-oxoglutarate (2-OG)-dependent dioxygenase, ... | | Authors: | Hou, X.D, Liu, X.Z, Yuan, Z.B, Rao, Y.J. | | Deposit date: | 2021-05-18 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Molecular Basis of the Unusual Seven-Membered Methylenedioxy Bridge Formation Catalyzed by Fe(II)/alpha-KG-Dependent Oxygenase CTB9
Acs Catalysis, 12, 2022
|
|
3QBT
 
 | | Crystal structure of OCRL1 540-678 in complex with Rab8a:GppNHp | | Descriptor: | Inositol polyphosphate 5-phosphatase OCRL-1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Hou, X, Hagemann, N, Schoebel, S, Blankenfeldt, W, Goody, R.S, Erdmann, K.S, Itzen, A. | | Deposit date: | 2011-01-14 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural basis for Lowe syndrome caused by mutations in the Rab-binding domain of OCRL1.
Embo J., 30, 2011
|
|
8HFK
 
 | |
8HFJ
 
 | | Crystal Structure of CbAR mutant (H162F) in complex with NADP+ and a bulky 1,3-cyclodiketone | | Descriptor: | 2-methyl-2-[(4-methylphenyl)methyl]cyclopentane-1,3-dione, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Versicolorin reductase | | Authors: | Hou, X.D, Yin, D.J, Rao, Y.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of an anthrol reductase inspires enantioselective synthesis of enantiopure hydroxycycloketones and beta-halohydrins.
Nat Commun, 14, 2023
|
|
2HNK
 
 | | Crystal structure of SAM-dependent O-methyltransferase from pathogenic bacterium Leptospira interrogans | | Descriptor: | DI(HYDROXYETHYL)ETHER, S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent O-methyltransferase, ... | | Authors: | Hou, X, Wei, Z, Gong, W. | | Deposit date: | 2006-07-13 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SAM-dependent O-methyltransferase from pathogenic bacterium Leptospira interrogans.
J.Struct.Biol., 159, 2007
|
|
