4LYR
 
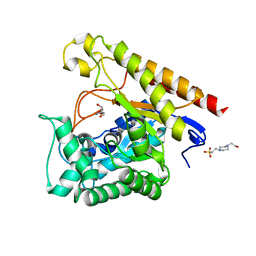 | | Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E301A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LYQ
 
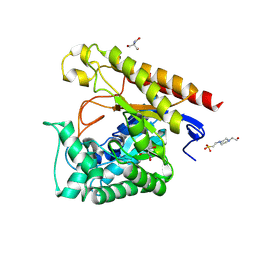 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E202A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase, ... | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7TAE
 
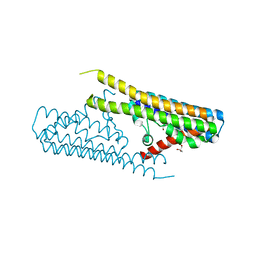 | |
8E4A
 
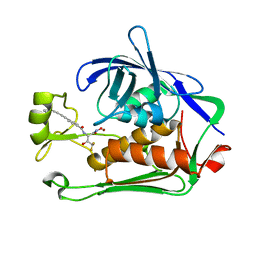 | | Pseudomonas LpxC in complex with LPC-233 | | Descriptor: | 4-(4-cyclopropylbuta-1,3-diyn-1-yl)-N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Najeeb, J, Zhou, P. | | Deposit date: | 2022-08-17 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.034 Å) | | Cite: | Preclinical safety and efficacy characterization of an LpxC inhibitor against Gram-negative pathogens.
Sci Transl Med, 15, 2023
|
|
6PIB
 
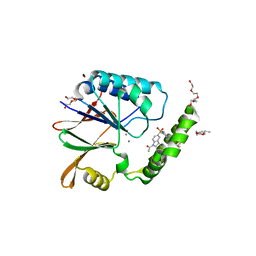 | | Structure of the Klebsiella pneumoniae LpxH-AZ1 complex | | Descriptor: | 1-[5-({4-[3-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-26 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PH9
 
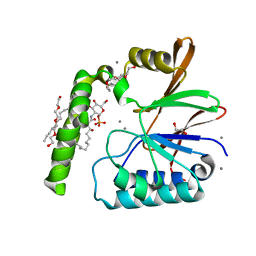 | | Crystal Structure of the Klebsiella pneumoniae LpxH-lipid X complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PJ3
 
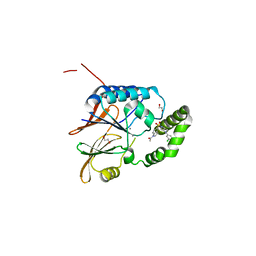 | | Crystal structure of the Klebsiella pneumoniae LpxH/JH-LPH-33 complex | | Descriptor: | 1,2-ETHANEDIOL, 1-[5-({4-[3-chloro-5-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-27 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7MK2
 
 | | CryoEM Structure of NPR1 | | Descriptor: | Regulatory protein NPR1, ZINC ION | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M, Bartesaghi, A, Zhou, P. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7MK3
 
 | | Crystal structure of NPR1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Regulatory protein NPR1, ... | | Authors: | Cheng, J, Wu, Q, Zhou, P. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
2VH0
 
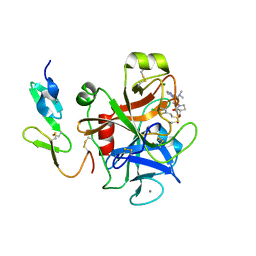 | | Structure and property based design of factor Xa inhibitors:biaryl pyrrolidin-2-ones incorporating basic heterocyclic motifs | | Descriptor: | 2-(5-chlorothiophen-2-yl)-N-[(3S)-1-(4-{2-[(dimethylamino)methyl]-1H-imidazol-1-yl}-2-fluorophenyl)-2-oxopyrrolidin-3-yl]ethanesulfonamide, ACTIVATED FACTOR XA HEAVY CHAIN, ACTIVATED FACTOR XA LIGHT CHAIN, ... | | Authors: | Young, R.J, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chan, C, Charbaut, M, Convery, M.A, Diallo, H, Hortense, E, Irving, W.R, Kelly, H.A, King, N.P, Kleanthous, S, Mason, A.M, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Zhou, P. | | Deposit date: | 2007-11-16 | | Release date: | 2008-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and property based design of factor Xa inhibitors: biaryl pyrrolidin-2-ones incorporating basic heterocyclic motifs.
Bioorg. Med. Chem. Lett., 18, 2008
|
|
7SS6
 
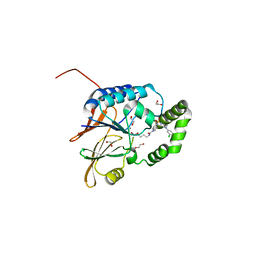 | | Structure of Klebsiella LpxH in complex with JH-LPH-45 | | Descriptor: | 1,2-ETHANEDIOL, 5-{4-[3-chloro-5-(trifluoromethyl)phenyl]piperazine-1-sulfonyl}-N-[5-(hydroxyamino)-5-oxopentyl]-2,3-dihydro-1H-indole-1-carboxamide, MANGANESE (II) ION, ... | | Authors: | Cho, J, Cochrane, C.S, Zhou, P. | | Deposit date: | 2021-11-09 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Development of LpxH Inhibitors Chelating the Active Site Dimanganese Metal Cluster of LpxH.
Chemmedchem, 18, 2023
|
|
7SS7
 
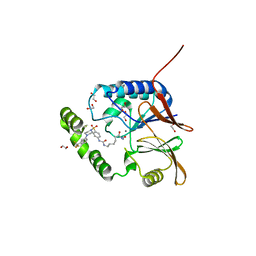 | | Crystal structure of Klebsiella LpxH in complex with JH-LPH-50 | | Descriptor: | 1,2-ETHANEDIOL, 6-{[(4-{4-[3-chloro-5-(trifluoromethyl)phenyl]piperazine-1-sulfonyl}phenyl)carbamoyl]amino}-N-hydroxyhexanamide, MANGANESE (II) ION, ... | | Authors: | Cho, J, Cochrane, C.S, Zhou, P. | | Deposit date: | 2021-11-09 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Development of LpxH Inhibitors Chelating the Active Site Dimanganese Metal Cluster of LpxH.
Chemmedchem, 18, 2023
|
|
9BD3
 
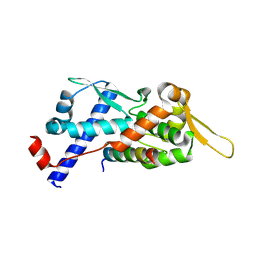 | | Structure of the MAGEA4 MHD-RAD18 R6BD Complex | | Descriptor: | E3 ubiquitin-protein ligase RAD18, Melanoma antigen A 4 | | Authors: | Forker, K, Zhou, P. | | Deposit date: | 2024-04-10 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of MAGEA4 MHD-RAD18 R6BD reveals a flipped binding mode compared to AlphaFold2 prediction.
Embo J., 43, 2024
|
|
6XQV
 
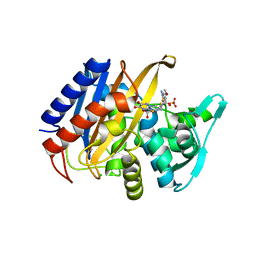 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae in a pre-acylation complex with ceftriaxone | | Descriptor: | CHLORIDE ION, Ceftriaxone, Probable peptidoglycan D,D-transpeptidase PenA, ... | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6XQZ
 
 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidoglycan D,D-transpeptidase PenA, ... | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6XQY
 
 | |
6XQX
 
 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae with the H514A mutation at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, Probable peptidoglycan D,D-transpeptidase PenA, SULFATE ION | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6WII
 
 | | Crystal structure of the K. pneumoniae LpxH/JH-LPH-41 complex | | Descriptor: | 5-({[4-({4-[3-chloro-5-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)phenyl]carbamoyl}amino)-N-hydroxypentanamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Cochrane, C.S, Cho, J, Fenton, B.A, Zhou, P. | | Deposit date: | 2020-04-09 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and evaluation of sulfonyl piperazine LpxH inhibitors.
Bioorg.Chem., 102, 2020
|
|
7OX9
 
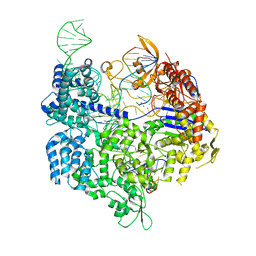 | | Target-bound SpCas9 complex with AAVS1 all-RNA guide | | Descriptor: | AAVS1 non-target DNA strand, AAVS1 target DNA strand, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Pacesa, M, Donohoue, P, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
6C59
 
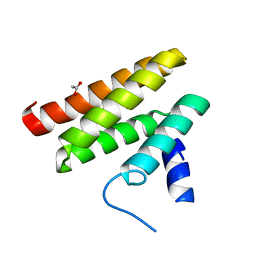 | | Chimeric Pol kappa RIR Rev1 C-terminal domain | | Descriptor: | ACETATE ION, Chimeric Pol kappa RIR Rev1 C-terminal domain | | Authors: | Wojtaszek, J.L, Zhou, P. | | Deposit date: | 2018-01-15 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Small Molecule Targeting Mutagenic Translesion Synthesis Improves Chemotherapy.
Cell, 178, 2019
|
|
6C8C
 
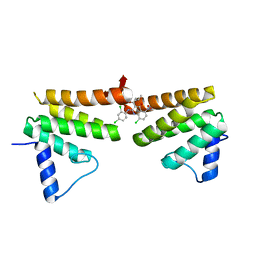 | | Chimeric Pol kappa RIR Rev1 C-terminal domain in complex with JHRE06 | | Descriptor: | 8-chloro-2-[(2,4-dichlorophenyl)amino]-3-(3-methylbutanoyl)-5-nitroquinolin-4(1H)-one, Chimeric protein of the Pol Kappa RIR helix and the Rev1 C-terminal domain | | Authors: | Najeeb, J, Zhou, P. | | Deposit date: | 2018-01-24 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Small Molecule Targeting Mutagenic Translesion Synthesis Improves Chemotherapy.
Cell, 178, 2019
|
|
6NE8
 
 | |
2J94
 
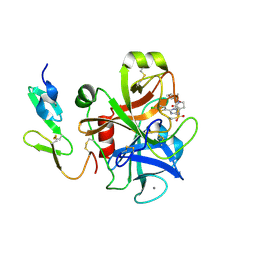 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 5-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-1H-1,2,4-TRIAZOLE-3-SULFONAMIDE, CALCIUM ION, COAGULATION FACTOR X | | Authors: | Chan, C, Borthwick, A.D, Brown, D, Campbell, M, Chaudry, L, Chung, C.W, Convery, M.A, Hamblin, J.N, Johnstone, L, Kelly, H.A, Kleanthous, S, Burns-Kurtis, C.L, Patikis, A, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Whitworth, C, Young, R.J, Zhou, P. | | Deposit date: | 2006-11-02 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Factor Xa Inhibitors: S1 Binding Interactions of a Series of N-{(3S)-1-[(1S)-1-Methyl-2-Morpholin-4-Yl-2-Oxoethyl]-2-Oxopyrrolidin-3-Yl}Sulfonamides.
J.Med.Chem., 50, 2007
|
|
2J4I
 
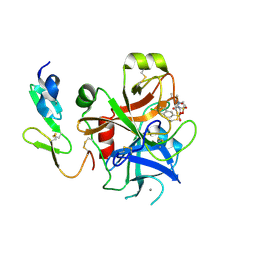 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 1-PYRROLIDINEACETAMIDE, 3-[[(6-CHLORO-2-NAPHTHALENYL)SULFONYL]AMINO]-ALPHA-METHYL-N-(1-METHYLETHYL)-N-[2-[(METHYLSULFONYL)AMINO]ETHYL]-2-OXO-, (ALPHAS,3S)-, ... | | Authors: | Young, R.J, Campbell, M, Borthwick, A.D, Brown, D, Chan, C, Convery, M.A, Crowe, M.C, Dayal, S, Diallo, H, Kelly, H.A, Paul King, N, Kleanthous, S, Kurtis, C.L, Mason, A.M, Mordaunt, J.E, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Smith, P.W, Watson, N.S, Weston, H.E, Zhou, P. | | Deposit date: | 2006-08-31 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure- and Property-Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Acyclic Alanyl Amides as P4 Motifs.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8DTG
 
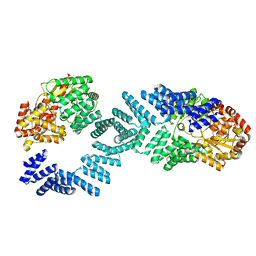 | | Cryo-EM structure of Arabidopsis SPY alternative conformation 1 | | Descriptor: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
