2A7O
 
 | | Solution Structure of the hSet2/HYPB SRI domain | | Descriptor: | Huntingtin interacting protein B | | Authors: | Li, M, Phatnani, H.P, Guan, Z, Sage, H, Greenleaf, A, Zhou, P. | | Deposit date: | 2005-07-05 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Set2 Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
7SS6
 
 | | Structure of Klebsiella LpxH in complex with JH-LPH-45 | | Descriptor: | 1,2-ETHANEDIOL, 5-{4-[3-chloro-5-(trifluoromethyl)phenyl]piperazine-1-sulfonyl}-N-[5-(hydroxyamino)-5-oxopentyl]-2,3-dihydro-1H-indole-1-carboxamide, MANGANESE (II) ION, ... | | Authors: | Cho, J, Cochrane, C.S, Zhou, P. | | Deposit date: | 2021-11-09 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Development of LpxH Inhibitors Chelating the Active Site Dimanganese Metal Cluster of LpxH.
Chemmedchem, 18, 2023
|
|
7SS7
 
 | | Crystal structure of Klebsiella LpxH in complex with JH-LPH-50 | | Descriptor: | 1,2-ETHANEDIOL, 6-{[(4-{4-[3-chloro-5-(trifluoromethyl)phenyl]piperazine-1-sulfonyl}phenyl)carbamoyl]amino}-N-hydroxyhexanamide, MANGANESE (II) ION, ... | | Authors: | Cho, J, Cochrane, C.S, Zhou, P. | | Deposit date: | 2021-11-09 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Development of LpxH Inhibitors Chelating the Active Site Dimanganese Metal Cluster of LpxH.
Chemmedchem, 18, 2023
|
|
5DRO
 
 | | Structure of the Aquifex aeolicus LpxC/LPC-011 Complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Lee, C.-J, Najeeb, J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
5DRP
 
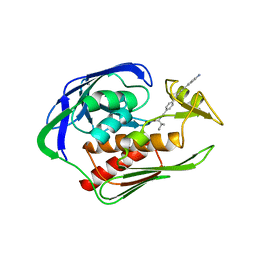 | | Structure of the AaLpxC/LPC-023 Complex | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, N~2~-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N-hydroxy-L-isoleucinamide, ... | | Authors: | Najeeb, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
4MQY
 
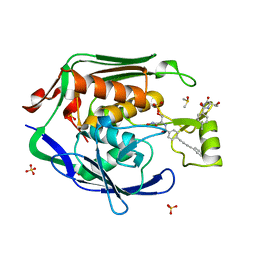 | | Crystal Structure of the Escherichia coli LpxC/LPC-138 complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-2-methyl-1-nitroso-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Lee, C.-J, Najeeb, J, Zhou, P. | | Deposit date: | 2013-09-17 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural Basis of the Promiscuous Inhibitor Susceptibility of Escherichia coli LpxC.
Acs Chem.Biol., 9, 2014
|
|
2UWP
 
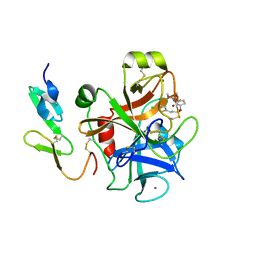 | | Factor Xa inhibitor complex | | Descriptor: | 2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}ETHANESULFONAMIDE, CALCIUM ION, COAGULATION FACTOR X, ... | | Authors: | Young, R.J, Brown, D, Burns-Kurtis, C.L, Chan, C, Convery, M.A, Hubbard, J.A, Kelly, H.A, Pateman, A.J, Patikis, A, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Zhou, P, Thorpe, J.H. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4K35
 
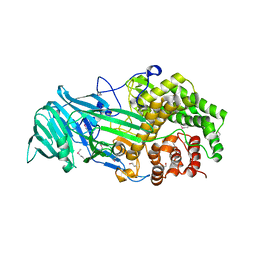 | | The structure of a glycoside hydrolase family 81 endo-[beta]-1,3-glucanase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, glycoside hydrolase family 81 endo-beta-1,3-glucanase | | Authors: | Jiang, Z.Q, Zhou, P, Chen, Z.Z, Yan, Q.J, Yang, S.Q, Hilgenfeld, R. | | Deposit date: | 2013-04-10 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | The structure of a glycoside hydrolase family 81
endo-[beta]-1,3-glucanase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2UWO
 
 | | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa | | Descriptor: | (2R)-2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}PROPENE-1-SULFONAMIDE, COAGULATION FACTOR X | | Authors: | Young, R.J, Brown, D, Burns-Kurtis, C.L, Chan, C, Convery, M.A, Hubbard, J.A, Kelly, H.A, Pateman, A.J, Patikis, A, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Zhou, P. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2UWL
 
 | | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa | | Descriptor: | 2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}ETHENESULFONAMIDE, COAGULATION FACTOR X | | Authors: | Young, R.J, Brown, D, Burns-Kurtis, C.L, Chan, C, Convery, M.A, Hubbard, J.A, Kelly, H.A, Pateman, A.J, Patikis, A, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Zhou, P. | | Deposit date: | 2007-03-22 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4J72
 
 | | Crystal Structure of polyprenyl-phosphate N-acetyl hexosamine 1-phosphate transferase | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Lee, S.Y, Chung, B.C, Gillespie, R.A, Kwon, D.Y, Guan, Z, Zhou, P, Hong, J. | | Deposit date: | 2013-02-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis.
Science, 341, 2013
|
|
4K3A
 
 | | The structure of a glycoside hydrolase family 81 endo-[beta]-1,3-glucanase | | Descriptor: | SULFATE ION, glycoside hydrolase family 81 endo-beta-1,3-glucanase | | Authors: | Jiang, Z.Q, Zhou, P, Chen, Z.Z, Yan, Q.J, Yang, S.Q, Hilgenfeld, R. | | Deposit date: | 2013-04-10 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a glycoside hydrolase family 81
endo-[beta]-1,3-glucanase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6AZR
 
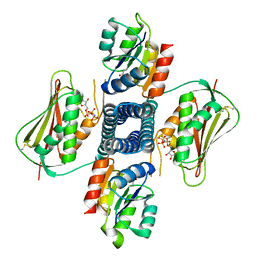 | | Crystal structure of the T264A HK853cp-BeF3-RR468 complex | | Descriptor: | Chemotaxis regulator-transmits chemoreceptor signals to flagelllar motor components CheY, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Rose, J, Zhou, P. | | Deposit date: | 2017-09-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.628 Å) | | Cite: | A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases.
Nat Commun, 8, 2017
|
|
1U8B
 
 | | Crystal structure of the methylated N-ADA/DNA complex | | Descriptor: | 5'-D(*AP*AP*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*T)-3', 5'-D(*TP*AP*AP*AP*TP*T)-3', 5'-D(P*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*AP*T)-3', ... | | Authors: | He, C, Hus, J.-C, Sun, L.J, Zhou, P, Norman, D.P.G, Dotsch, V, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2004-08-05 | | Release date: | 2005-10-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A methylation-dependent electrostatic switch controls DNA repair and transcriptional activation by E. coli ada.
Mol.Cell, 20, 2005
|
|
4LYP
 
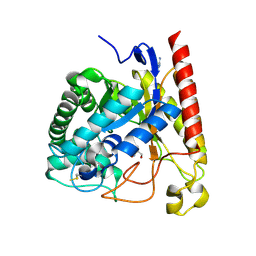 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Exo-beta-1,4-mannosidase, GUANIDINE | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1ZGW
 
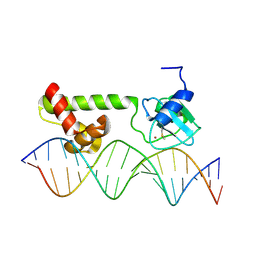 | | NMR structure of E. Coli Ada protein in complex with DNA | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*TP*TP*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*A)-3', 5'-D(*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*TP*AP*AP*TP*TP*TP*GP*C)-3', Ada polyprotein, ... | | Authors: | He, C, Hus, J.C, Sun, L.J, Zhou, P, Norman, D.P, Doetsch, V, Wei, H, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2005-04-22 | | Release date: | 2005-10-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A Methylation-Dependent Electrostatic Switch Controls DNA Repair and Transcriptional Activation by E. coli Ada.
Mol.Cell, 20, 2005
|
|
5XBZ
 
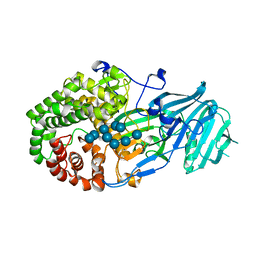 | | Crystal structure of GH family 81 beta-1,3-glucanase from Rhizomucr miehei complexed with laminaripentaose | | Descriptor: | Endo-beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Yang, S, Qin, Z, Zhou, P, Yan, Q, Jiang, Z. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of glycoside hydrolase family 81 beta-1,3-glucanase
To Be Published
|
|
5U86
 
 | | Structure of the Aquifex aeolicus LpxC/LPC-069 complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]-4-({4-[(morpholin-4-yl)methyl]phenyl}ethynyl)benzamide, ... | | Authors: | Najeeb, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2016-12-13 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Curative Treatment of Severe Gram-Negative Bacterial Infections by a New Class of Antibiotics Targeting LpxC.
MBio, 8, 2017
|
|
3P3G
 
 | | Crystal Structure of the Escherichia coli LpxC/LPC-009 complex | | Descriptor: | 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, N-[(1S,2R)-2-hydroxy-1-(hydroxycarbamoyl)propyl]-4-(4-phenylbuta-1,3-diyn-1-yl)benzamide, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2010-10-04 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Species-specific and inhibitor-dependent conformations of LpxC: implications for antibiotic design.
Chem.Biol., 18, 2011
|
|
4NRR
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannosyl-fructose | | Descriptor: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-beta-D-fructofuranose | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-11-27 | | Release date: | 2014-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3PS2
 
 | | Crystal structure of the Escherichia Coli LPXC/LPC-012 complex | | Descriptor: | 4-[4-(3-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-nitroso-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Syntheses, structures and antibiotic activities of LpxC inhibitors based on the diacetylene scaffold.
Bioorg.Med.Chem., 19, 2011
|
|
3QC1
 
 | | Protein Phosphatase Subunit: Alpha4 | | Descriptor: | Immunoglobulin-binding protein 1 | | Authors: | Spiller, B.W, LeNoue-Newton, M.L, Watkins, G.R, Germane, K.L, Zhou, P, McCorvey, L.R, Wadzinski, B.E. | | Deposit date: | 2011-01-14 | | Release date: | 2011-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Mid1 and PP2Ac binding domains of Alpha4 are both required for Alpha4 to inhibit PP2Ac degradation
TO BE PUBLISHED
|
|
4NRS
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose | | Descriptor: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-11-27 | | Release date: | 2014-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3PS1
 
 | | Crystal structure of the Escherichia Coli LPXC/LPC-011 complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Syntheses, structures and antibiotic activities of LpxC inhibitors based on the diacetylene scaffold.
Bioorg.Med.Chem., 19, 2011
|
|
3PS3
 
 | | Crystal structure of the Escherichia Coli LPXC/LPC-053 complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3S)-3-hydroxy-1-nitroso-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Syntheses, structures and antibiotic activities of LpxC inhibitors based on the diacetylene scaffold.
Bioorg.Med.Chem., 19, 2011
|
|
