5ZV2
 
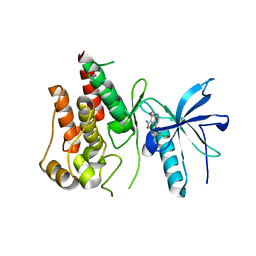 | | FGFR-1 in complex with ligand lenvatinib | | Descriptor: | 4-{3-chloro-4-[(cyclopropylcarbamoyl)amino]phenoxy}-7-methoxyquinoline-6-carboxamide, Fibroblast growth factor receptor 1 | | Authors: | Matsuki, M, Hoshi, T, Yamamoto, Y, Ikemori-Kawada, M, Minoshima, Y, Funahashi, Y, Matsui, J. | | Deposit date: | 2018-05-09 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Lenvatinib inhibits angiogenesis and tumor fibroblast growth factor signaling pathways in human hepatocellular carcinoma models.
Cancer Med, 7, 2018
|
|
8Z5L
 
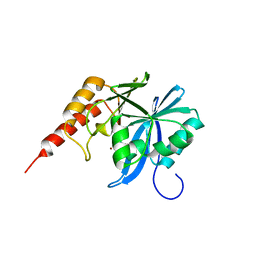 | | Crystal structure of metallo-beta-lactamse, IMP-1, complexed with a quinolinone-based inhibitor | | Descriptor: | 3-[2-azanyl-5-[2-cyclohexylethyl-[3-(4-methylphenoxy)propyl]amino]phenyl]propanoic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Kamo, T, Kuroda, K, Nimura, S, Guo, Y, Kondo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Development of Inhibitory Compounds for Metallo-beta-lactamase through Computational Design and Crystallographic Analysis.
Biochemistry, 63, 2024
|
|
8JYH
 
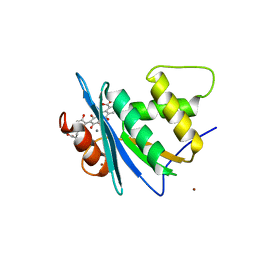 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid C | | Descriptor: | 7-[5-[(2~{S})-2-azanyl-3-oxidanyl-3-oxidanylidene-propyl]-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,HIV-1 Reverse Transcriptase RNase H active domain, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
8JYJ
 
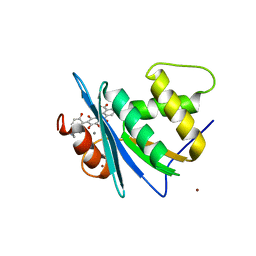 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid A | | Descriptor: | 7-[5-(2-acetamidoethyl)-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,HIV-1 Reverse Transcriptase RNase H active domain, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
8JYI
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid E | | Descriptor: | 7-[5-(2-azanylethyl)-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,Ribonuclease H, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
4KT8
 
 | | The complex structure of Rv3378c-Y51FY90F with substrate, TPP | | Descriptor: | (2E)-3-methyl-5-[(1R,2S,8aS)-1,2,5,5-tetramethyl-1,2,3,5,6,7,8,8a-octahydronaphthalen-1-yl]pent-2-en-1-yl trihydrogen diphosphate, Diterpene synthase, PHOSPHATE ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
5JHT
 
 | | Apo form of influenza strain H1N1 polymerase acidic subunit N-terminal region crystallized with potassium sodium tartrate | | Descriptor: | L(+)-TARTARIC ACID, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Hoshino, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Influence of Precipitants on Molecular Arrangements and Space Groups of Protein Crystals
Cryst.Growth Des., 17, 2017
|
|
2RQH
 
 | | Structure of GSPT1/ERF3A-PABC | | Descriptor: | G1 to S phase transition 1, Polyadenylate-binding protein 1 | | Authors: | Osawa, M, Nakanishi, T, Hosoda, N, Uchida, S, Hoshino, T, Katada, I, Shimada, I. | | Deposit date: | 2009-05-08 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic Translation Termination Factor Gspt/Erf3 Recognizes Pabp with Chemical Exchange Using Two Overlapping Motifs
To be Published
|
|
2RQG
 
 | | Structure of GSPT1/ERF3A-PABC | | Descriptor: | G1 to S phase transition 1, Polyadenylate-binding protein 1 | | Authors: | Osawa, M, Nakanishi, T, Hosoda, N, Uchida, S, Hoshino, T, Katada, I, Shimada, I. | | Deposit date: | 2009-05-08 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic Translation Termination Factor Gspt/Erf3 Recognizes Pabp with Chemical Exchange Using Two Overlapping Motifs
To be Published
|
|
5JHV
 
 | | Apo form of influenza strain H1N1 polymerase acidic subunit N-terminal region crystallized with polyethylene glycol 8000 | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Hoshino, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.749 Å) | | Cite: | Influence of Precipitants on Molecular Arrangements and Space Groups of Protein Crystals
Cryst.Growth Des., 17, 2017
|
|
6JKB
 
 | | Crystal structure of metallo-beta-lactamse, NDM-1, in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Kamo, T, Kuroda, K, Kondo, S, Hayashi, U, Fudo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Identification of the Inhibitory Compounds for Metallo-beta-lactamases and Structural Analysis of the Binding Modes.
Chem Pharm Bull (Tokyo), 69, 2021
|
|
6JKA
 
 | | Crystal structure of metallo-beta-lactamse, IMP-1, in complex with a thiazole-bearing inhibitor | | Descriptor: | 3-[2-azanyl-5-[2-(phenoxymethyl)-1,3-thiazol-4-yl]phenyl]propanoic acid, 6-[2-(phenoxymethyl)-1,3-thiazol-4-yl]-3,4-dihydro-1H-quinolin-2-one, Metallo-beta-lactamase type 2, ... | | Authors: | Kamo, T, Kuroda, K, Kondo, S, Hayashi, U, Fudo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Identification of the Inhibitory Compounds for Metallo-beta-lactamases and Structural Analysis of the Binding Modes.
Chem Pharm Bull (Tokyo), 69, 2021
|
|
6JE7
 
 | | Crystal structure of human serum albumin crystallized by ammonium sulfate | | Descriptor: | Serum albumin | | Authors: | Kitahara, M, Fudo, S, Yoneda, T, Nukaga, M, Hoshino, T. | | Deposit date: | 2019-02-04 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Anisotropic Distribution of Ammonium Sulfate Ions in Protein Crystallization
Cryst.Growth Des., 19, 2019
|
|
4ZHZ
 
 | | Endonuclease inhibitor bound to influenza strain H1N1 polymerase acidic subunit N-terminal region with expelling one of the metal ions in the active site | | Descriptor: | 5-(2-chlorobenzyl)-2-hydroxy-3-nitrobenzaldehyde, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Neya, S, Hoshino, T. | | Deposit date: | 2015-04-27 | | Release date: | 2015-05-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit
Biochemistry, 55, 2016
|
|
4YYL
 
 | | Phenolic acid derivative bound to influenza strain H1N1 polymerase subunit PA endonuclease | | Descriptor: | 2-(4-fluorophenoxy)-1-(2,3,4-trihydroxyphenyl)ethanone, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Neya, S, Hoshino, T. | | Deposit date: | 2015-03-24 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Structural and computational study on inhibitory compounds for endonuclease activity of influenza virus polymerase
Bioorg.Med.Chem., 23, 2015
|
|
5FDG
 
 | | Endonuclease inhibitor 3 bound to influenza strain H1N1 polymerase acidic subunit N-terminal region at pH 7.0 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Hoshino, T. | | Deposit date: | 2015-12-16 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit
Biochemistry, 55, 2016
|
|
5FDD
 
 | | Endonuclease inhibitor 1 bound to influenza strain H1N1 polymerase acidic subunit N-terminal region at pH 7.0 | | Descriptor: | 5-(2-chlorobenzyl)-2-hydroxy-3-nitrobenzaldehyde, MANGANESE (II) ION, Polymerase acidic protein,Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Hoshino, T. | | Deposit date: | 2015-12-16 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit
Biochemistry, 55, 2016
|
|
5I13
 
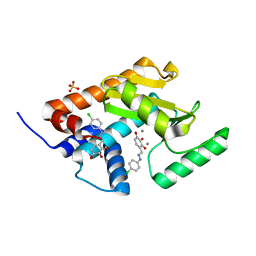 | | Endonuclease inhibitor 2 bound to influenza strain H1N1 polymerase acidic subunit N-terminal region at pH 7.0 | | Descriptor: | 4-{(E)-[2-(4-chlorophenyl)hydrazinylidene]methyl}benzene-1,2,3-triol, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Hoshino, T. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit
Biochemistry, 55, 2016
|
|
4ZI0
 
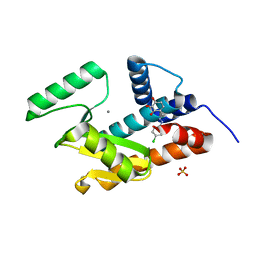 | | Endonuclease inhibitor bound to influenza strain H1N1 polymerase acidic subunit N-terminal region without a chelation to the metal ions in the active site | | Descriptor: | 4-{(E)-[2-(4-chlorophenyl)hydrazinylidene]methyl}benzene-1,2,3-triol, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Neya, S, Hoshino, T. | | Deposit date: | 2015-04-27 | | Release date: | 2015-05-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit
Biochemistry, 55, 2016
|
|
4ZQQ
 
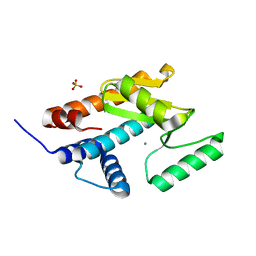 | | Apo form of influenza strain H1N1 polymerase acidic subunit N-terminal region | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein,Polymerase acidic protein, SULFATE ION | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Neya, S, Hoshino, T. | | Deposit date: | 2015-05-11 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit.
Biochemistry, 55, 2016
|
|
3NLA
 
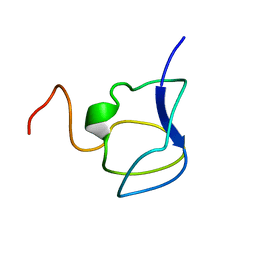 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, 40 STRUCTURES | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
5ZA2
 
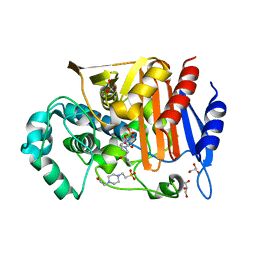 | | Fox-4 beta-lactamase complexed with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase, ... | | Authors: | Nukaga, M, Hoshino, T, Papp-Wallace, K.M, Bonomo, R.A. | | Deposit date: | 2018-02-06 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Probing the Mechanism of Inactivation of the FOX-4 Cephamycinase by Avibactam
Antimicrob. Agents Chemother., 62, 2018
|
|
3RDN
 
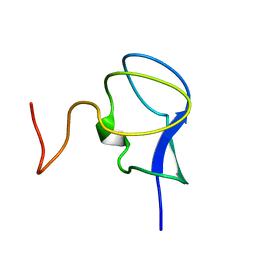 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
5ZXW
 
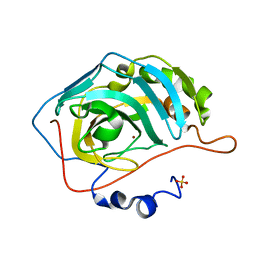 | | Crystal structure of human carbonic anhydrase II crystallized by ammonium sulfate | | Descriptor: | Carbonic anhydrase 2, SULFATE ION, ZINC ION | | Authors: | Kitahara, M, Fudo, S, Yoneda, T, Nukaga, M, Hoshino, T. | | Deposit date: | 2018-05-22 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.316 Å) | | Cite: | Anisotropic Distribution of Ammonium Sulfate Ions in Protein Crystallization
Cryst.Growth Des., 19, 2019
|
|
5ZZE
 
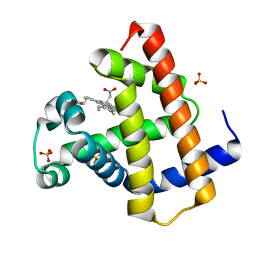 | | Crystal structure of horse myoglobin crystallized by ammonium sulfate | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kitahara, M, Fudo, S, Yoneda, T, Nukaga, M, Hoshino, T. | | Deposit date: | 2018-06-01 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | Anisotropic Distribution of Ammonium Sulfate Ions in Protein Crystallization
Cryst.Growth Des., 19, 2019
|
|
