1H8O
 
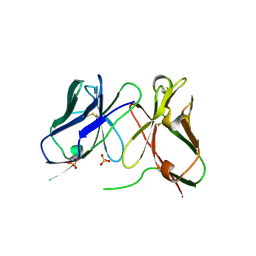 | | Three-dimensional structure of anti-ampicillin single chain Fv fragment. | | Descriptor: | MUTANT AL2 6E7P9G, SULFATE ION | | Authors: | Burmester, J, Spinelli, S, Pugliese, L, Krebber, A, Honegger, A, Jung, S, Schimmele, B, Cambillau, C, Pluckthun, A. | | Deposit date: | 2001-02-14 | | Release date: | 2001-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Selection, Characterization and X-Ray Structure of Anti-Ampicillin Single-Chain Fv Fragments from Phage-Displayed Murine Antibody Libraries
J.Mol.Biol., 309, 2001
|
|
1H8N
 
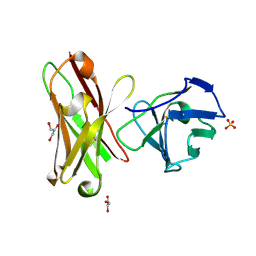 | | Three-dimensional structure of anti-ampicillin single chain Fv fragment from phage-displayed murine antibody libraries | | Descriptor: | GLYCEROL, MUTANT AL2 6E7S9G, SULFATE ION | | Authors: | Jung, S, Spinelli, S, Schimmele, B, Honegger, A, Pugliese, L, Cambillau, C, Pluckthun, A. | | Deposit date: | 2001-02-14 | | Release date: | 2001-08-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The Importance of Framework Residues H6, H7 and H10 in Antibody Heavy Chains: Experimental Evidence for a New Structural Subclassification of Antibody V(H) Domains
J.Mol.Biol., 309, 2001
|
|
5FIO
 
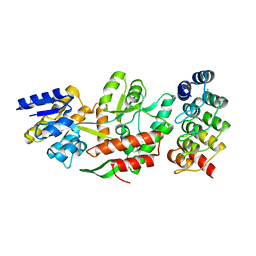 | | DARPins as a new tool for experimental phasing in protein crystallography | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, MERCURY (II) ION, NI3C DARPIN MUTANT5 HG-SITE N1 | | Authors: | Batyuk, A, Honegger, A, Andres, F, Briand, C, Gruetter, M, Plueckthun, A. | | Deposit date: | 2015-09-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Darpins as a New Tool for Experimental Phasing in Protein Crystallography
To be Published
|
|
6ZQK
 
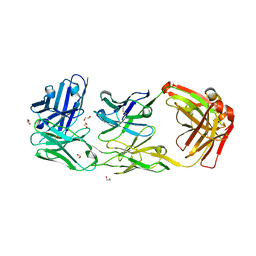 | | HER2-binding scFv-Fab fusion 841 | | Descriptor: | 1,2-ETHANEDIOL, 841 heavy chain, 841 light chain | | Authors: | Kast, F, Schwill, M, Stueber, J.C, Pfundstein, S, Nagy-Davidescu, G, Monne Rodriguez, J.M, Seehusen, F, Richter, C.P, Honegger, A, Hartmann, K.P, Weber, T.G, Kroener, F, Ernst, P, Piehler, J, Plueckthun, A. | | Deposit date: | 2020-07-09 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engineering an anti-HER2 biparatopic antibody with a multimodal mechanism of action.
Nat Commun, 12, 2021
|
|
5FIN
 
 | | DARPins as a new tool for experimental phasing in protein crystallography | | Descriptor: | MERCURY (II) ION, NI3C DARPIN MUTANT5 HG-SITE N1 | | Authors: | Batyuk, A, Honegger, A, Andres, F, Briand, C, Gruetter, M, Plueckthun, A. | | Deposit date: | 2015-09-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Darpins as a New Tool for Experimental Phasing in Protein Crystallography
To be Published
|
|
1C12
 
 | | INSIGHT IN ODORANT PERCEPTION: THE CRYSTAL STRUCTURE AND BINDING CHARACTERISTICS OF ANTIBODY FRAGMENTS DIRECTED AGAINST THE MUSK ODORANT TRASEOLIDE | | Descriptor: | PROTEIN (ANTIBODY FRAGMENT FAB), TRAZEOLIDE | | Authors: | Langedijk, A.C, Spinelli, S, Anguille, C, Hermans, P, Nederlof, J, Butenandt, J, Honegger, A, Cambillau, C, Pluckthun, A. | | Deposit date: | 1999-07-20 | | Release date: | 1999-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into odorant perception: the crystal structure and binding characteristics of antibody fragments directed against the musk odorant traseolide.
J.Mol.Biol., 292, 1999
|
|
5OGI
 
 | | Complex of a binding protein and human adenovirus C 5 hexon | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Schmid, M, Ernst, P, Honegger, A, Suomalainen, M, Zimmermann, M, Braun, L, Stauffer, S, Thom, C, Dreier, B, Eibauer, M, Kipar, A, Vogel, V, Greber, U.F, Medalia, O, Plueckthun, A. | | Deposit date: | 2017-07-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Adenoviral vector with shield and adapter increases tumor specificity and escapes liver and immune control.
Nat Commun, 9, 2018
|
|
1KTR
 
 | | Crystal Structure of the Anti-His Tag Antibody 3D5 Single-Chain Fragment (scFv) in Complex with a Oligohistidine peptide | | Descriptor: | Anti-his tag antibody 3d5 variable light chain, Peptide linker, Anti-his tag antibody 3d5 variable heavy chain, ... | | Authors: | Kaufmann, M, Lindner, P, Honegger, A, Blank, K, Tschopp, M, Capitani, G, Plueckthun, A, Gruetter, M.G. | | Deposit date: | 2002-01-17 | | Release date: | 2002-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the anti-His tag antibody 3D5 single-chain fragment complexed to its antigen.
J.Mol.Biol., 318, 2002
|
|
6F5E
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_10_47 in complex JNK1a1 and JIP1 peptide | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 1, DD_D12_10_47, Mitogen-activated protein kinase 8 | | Authors: | Wu, Y, Mittl, P.R, Honegger, A, Batyuk, A, Plueckthun, A. | | Deposit date: | 2017-12-01 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_10_47 in complex JNK1a1 and JIP1 peptide
To be published
|
|
6EQC
 
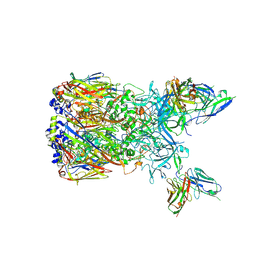 | | Cryo-EM reconstruction of a complex of a binding protein and human adenovirus C5 hexon | | Descriptor: | Hexon protein, scFv of 9C12 antibody | | Authors: | Schmid, M, Ernst, P, Honegger, A, Suomalainen, M, Zimmermann, M, Braun, L, Stauffer, S, Thom, C, Dreier, B, Eibauer, M, Kipar, A, Vogel, V, Greber, U.F, Medalia, O, Plueckthun, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Adenoviral vector with shield and adapter increases tumor specificity and escapes liver and immune control.
Nat Commun, 9, 2018
|
|
1H8S
 
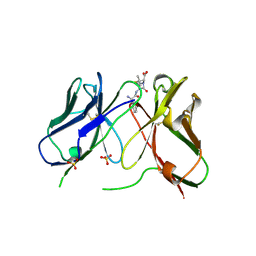 | | Three-dimensional structure of anti-ampicillin single chain Fv fragment complexed with the hapten. | | Descriptor: | (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, MUTANT AL2 6E7P9G, SULFATE ION | | Authors: | Burmester, J, Spinelli, S, Pugliese, L, Krebber, A, Honegger, A, Jung, S, Schimmele, B, Cambillau, C, Pluckthun, A. | | Deposit date: | 2001-02-15 | | Release date: | 2001-08-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selection, Characterization and X-Ray Structure of Anti-Ampicillin Single-Chain Fv Fragments from Phage-Displayed Murine Antibody Libraries
J.Mol.Biol., 309, 2001
|
|
1I3G
 
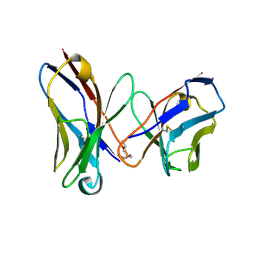 | | CRYSTAL STRUCTURE OF AN AMPICILLIN SINGLE CHAIN FV, FORM 1, FREE | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ANTIBODY FV FRAGMENT | | Authors: | Jung, S, Spinelli, S, Schimmele, B, Honegger, A, Pugliese, L, Cambillau, C, Pluckthun, A. | | Deposit date: | 2001-02-15 | | Release date: | 2001-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Selection, characterization and x-ray structure of anti-ampicillin single-chain Fv fragments from phage-displayed murine antibody libraries.
J.Mol.Biol., 309, 2001
|
|
2A9N
 
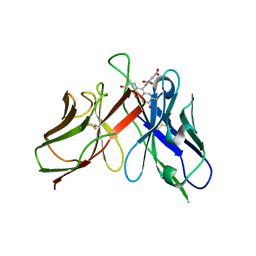 | | A Mutation Designed to Alter Crystal Packing Permits Structural Analysis of a Tight-binding Fluorescein-scFv complex | | Descriptor: | 4-(2,7-DIFLUORO-6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)ISOPHTHALIC ACID, fluorescein-scfv | | Authors: | Cambillau, C, Spinelli, S, Honegger, A, Pluckthun, A. | | Deposit date: | 2005-07-12 | | Release date: | 2005-10-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A mutation designed to alter crystal packing permits structural analysis of a tight-binding fluorescein-scFv complex.
Protein Sci., 14, 2005
|
|
2A9M
 
 | |
6SA7
 
 | | DARPin-Armadillo fusion C8long83 | | Descriptor: | DARPin-Armadillo fusion C8long83 | | Authors: | Ernst, P, Honegger, A, van der Valk, F, Ewald, C, Mittl, P.R.E, Plucktun, A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Rigid fusions of designed helical repeat binding proteins efficiently protect a binding surface from crystal contacts.
Sci Rep, 9, 2019
|
|
6SA8
 
 | | ring-like DARPin-Armadillo fusion H83_D01 | | Descriptor: | 1,2-ETHANEDIOL, LYS-ARG-LYS-ARG-LYS-ARG-LYS-ARG-LYS-ARG, ring-like DARPin-Armadillo fusion H83_D01 | | Authors: | Ernst, P, Honegger, A, van der Valk, F, Ewald, C, Mittl, P.R.E, Plucktun, A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rigid fusions of designed helical repeat binding proteins efficiently protect a binding surface from crystal contacts.
Sci Rep, 9, 2019
|
|
6SA6
 
 | | DARPin-Armadillo fusion A5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DARPin-Armadillo fusion A5 | | Authors: | Ernst, P, Honegger, A, van der Valk, F, Ewald, C, Mittl, P.R.E, Pluckthun, A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rigid fusions of designed helical repeat binding proteins efficiently protect a binding surface from crystal contacts.
Sci Rep, 9, 2019
|
|
4D49
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARGININE, ARMADILLO REPEAT PROTEIN ARM00027, POLY ARG DECAPEPTIDE | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4D4E
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARMADILLO REPEAT PROTEIN ARM00016, GLYCEROL | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
5AQ8
 
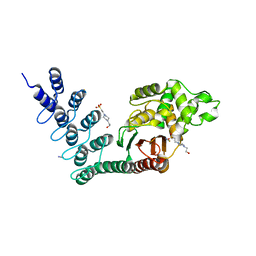 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, OFF7_DB12V4, THIOCYANATE ION | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
5AQB
 
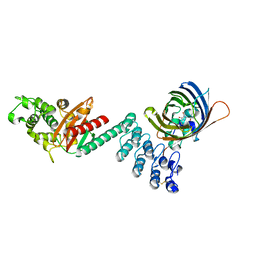 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | 3G61_DB15V4, GREEN FLUORESCENT PROTEIN | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
5AQ9
 
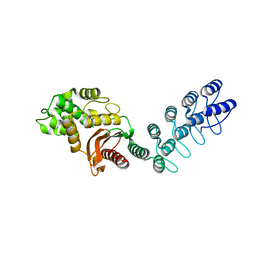 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, OFF7_DB08V4 | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
5AQA
 
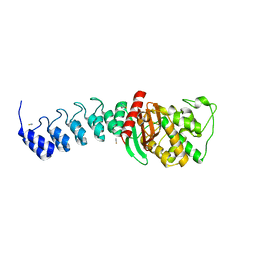 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | OFF7_DB04V3, THIOCYANATE ION | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
5AQ7
 
 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | D12_DB04V3, MALONATE ION | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
5LW1
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_232_11_D12 in complex JNK1a1 and JIP1 peptide | | Descriptor: | ADENOSINE, C-Jun-amino-terminal kinase-interacting protein 1, DD_232_11_D12, ... | | Authors: | Wu, Y, Batyuk, A, Mittl, P.R, Honegger, A, Plueckthun, A. | | Deposit date: | 2016-09-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for the Selective Inhibition of c-Jun N-Terminal Kinase 1 Determined by Rigid DARPin-DARPin Fusions.
J.Mol.Biol., 430, 2018
|
|
