3PLU
 
 | | Structure of Hub-1 protein in complex with Snu66 peptide (HINDI) | | Descriptor: | 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, Ubiquitin-like modifier HUB1 | | Authors: | Mishra, S.K, Ammon, T, Popowicz, G.M, Krajewski, M, Nagel, R.J, Ares, M, Holak, T.A, Jentsch, S. | | Deposit date: | 2010-11-15 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing.
Nature, 474, 2011
|
|
3PLV
 
 | | Structure of Hub-1 protein in complex with Snu66 peptide (HINDII) | | Descriptor: | 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, Ubiquitin-like modifier HUB1 | | Authors: | Mishra, S.K, Ammon, T, Popowicz, G.M, Krajewski, M, Nagel, R.J, Ares, M, Holak, T.A, Jentsch, S. | | Deposit date: | 2010-11-15 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing.
Nature, 474, 2011
|
|
3RMP
 
 | | Structural basis for the recognition of attP substrates by P4-like integrases | | Descriptor: | 5'-D(*TP*AP*AP*TP*GP*AP*CP*CP*AP*CP*CP*AP*AP*TP*A)-3', 5'-D(*TP*AP*TP*TP*GP*GP*TP*GP*GP*TP*CP*AP*TP*TP*A)-3', CP4-like integrase | | Authors: | Szwagierczak, A, Popowicz, G.M, Holak, T.A, Rakin, A, Antonenka, U. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for the recognition of attP substrates by P4-like integrases
To be Published
|
|
3TC5
 
 | | Selective targeting of disease-relevant protein binding domains by O-phosphorylated natural product derivatives | | Descriptor: | (11alpha,16alpha)-9-fluoro-11,17-dihydroxy-16-methyl-3,20-dioxopregna-1,4-dien-21-yl dihydrogen phosphate, HEXAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Graeber, M, Janczyk, W, Sperl, B, Elumalai, N, Kozany, C, Hausch, F, Holak, T.A, Berg, T. | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective targeting of disease-relevant protein binding domains by o-phosphorylated natural product derivatives.
Acs Chem.Biol., 6, 2011
|
|
3TJ2
 
 | | Structure of a novel submicromolar MDM2 inhibitor | | Descriptor: | 3-{(1S)-2-(tert-butylamino)-1-[(4-chlorobenzyl)(formyl)amino]-2-oxoethyl}-6-chloro-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, POTASSIUM ION | | Authors: | Wolf, S, Huang, Y, Popowicz, G.M, Goda, S, Holak, T.A, Doemling, A. | | Deposit date: | 2011-08-23 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ugi Multicomponent Reaction Derived p53-Mdm2 Antagonists
To be published
|
|
3LBL
 
 | | Structure of human MDM2 protein in complex with Mi-63-analog | | Descriptor: | (2'R,3R,4'R,5'R)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-N-(2-morpholin-4-ylethyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
3LBK
 
 | | Structure of human MDM2 protein in complex with a small molecule inhibitor | | Descriptor: | 6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
3LBJ
 
 | | Structure of human MDMX protein in complex with a small molecule inhibitor | | Descriptor: | N-[(3S)-1-({6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indol-2-yl}carbonyl)pyrrolidin-3-yl]-N,N',N'-trimethylpropane-1,3-diamine, Protein Mdm4, SULFATE ION | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
2DSQ
 
 | | Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor-binding protein 1, Insulin-like growth factor-binding protein 4 | | Authors: | Sitar, T, Popowicz, G.M, Siwanowicz, I, Huber, R, Holak, T.A. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3MN6
 
 | | Structures of actin-bound WH2 domains of Spire and the implication for filament nucleation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION, ... | | Authors: | Ducka, A.M, Sitar, T, Popowicz, G.M, Huber, R, Holak, T.A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MN9
 
 | | Structures of actin-bound WH2 domains of Spire and the implication for filament nucleation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION, ... | | Authors: | Ducka, A.M, Sitar, T, Popowicz, G.M, Huber, R, Holak, T.A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MN7
 
 | | Structures of actin-bound WH2 domains of Spire and the implication for filament nucleation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION, ... | | Authors: | Ducka, A.M, Sitar, T, Popowicz, G.M, Huber, R, Holak, T.A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MMV
 
 | | Structures of actin-bound WH2 domains of Spire and the implication for filament nucleation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION, ... | | Authors: | Ducka, A.M, Sitar, T, Popowicz, G.M, Huber, R, Holak, T.A. | | Deposit date: | 2010-04-20 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MN5
 
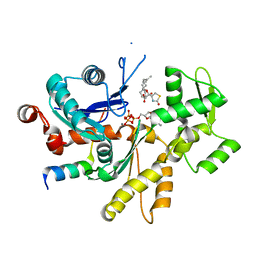 | | Structures of actin-bound WH2 domains of Spire and the implication for filament nucleation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Ducka, A.M, Sitar, T, Popowicz, G.M, Huber, R, Holak, T.A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2AS9
 
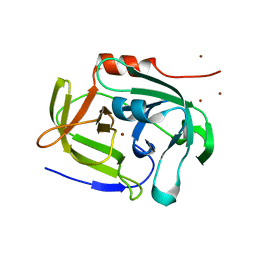 | | Functional and structural characterization of Spl proteases from staphylococcus aureus | | Descriptor: | ZINC ION, serine protease | | Authors: | Popowicz, G.M, Dubin, G, Stec-Niemczyk, J, Czarny, A, Dubin, A, Potempa, J, Holak, T.A. | | Deposit date: | 2005-08-23 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional and Structural Characterization of Spl Proteases from Staphylococcus aureus
J.Mol.Biol., 358, 2006
|
|
2DSP
 
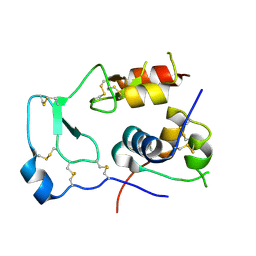 | | Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor-binding protein 4 | | Authors: | Sitar, T, Popowicz, G.M, Siwanowicz, I, Huber, R, Holak, T.A. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DSR
 
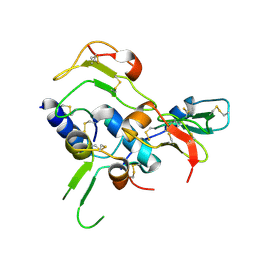 | | Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor-binding protein 4 | | Authors: | Sitar, T, Popowicz, G.M, Siwanowicz, I, Huber, R, Holak, T.A. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1WQJ
 
 | | Structural Basis for the Regulation of Insulin-Like Growth Factors (IGFs) by IGF Binding Proteins (IGFBPs) | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor binding protein 4 | | Authors: | Siwanowicz, I, Popowicz, G.M, Wisniewska, M, Huber, R, Kuenkele, K.P, Lang, K, Engh, R.A, Holak, T.A. | | Deposit date: | 2004-09-29 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the regulation of insulin-like growth factors by IGF binding proteins
Structure, 13, 2005
|
|
1ZLH
 
 | | Crystal structure of the tick carboxypeptidase inhibitor in complex with bovine carboxypeptidase A | | Descriptor: | Carboxypeptidase A1, ZINC ION, carboxypeptidase inhibitor | | Authors: | Arolas, J.L, Popowicz, G.M, Lorenzo, J, Sommerhoff, C.P, Huber, R, Aviles, F.X, Holak, T.A. | | Deposit date: | 2005-05-06 | | Release date: | 2005-07-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Three-Dimensional Structures of Tick Carboxypeptidase Inhibitor in Complex with A/B Carboxypeptidases Reveal a Novel Double-headed Binding Mode
J.Mol.Biol., 350, 2005
|
|
1YZ5
 
 | |
1ZLI
 
 | | Crystal structure of the tick carboxypeptidase inhibitor in complex with human carboxypeptidase B | | Descriptor: | Carboxypeptidase B, ZINC ION, carboxypeptidase inhibitor | | Authors: | Arolas, J.L, Popowicz, G.M, Lorenzo, J, Sommerhoff, C.P, Huber, R, Aviles, F.X, Holak, T.A. | | Deposit date: | 2005-05-06 | | Release date: | 2005-07-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Three-Dimensional Structures of Tick Carboxypeptidase Inhibitor in Complex with A/B Carboxypeptidases Reveal a Novel Double-headed Binding Mode
J.Mol.Biol., 350, 2005
|
|
1I16
 
 | | STRUCTURE OF INTERLEUKIN 16: IMPLICATIONS FOR FUNCTION, NMR, 20 STRUCTURES | | Descriptor: | INTERLEUKIN 16 | | Authors: | Muehlhahn, P, Zweckstetter, M, Georgescu, J, Ciosto, C, Renner, C, Lanzendoerfer, M, Lang, K, Ambrosius, D, Baier, M, Kurth, R, Holak, T.A. | | Deposit date: | 1998-05-20 | | Release date: | 1999-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of interleukin 16 resembles a PDZ domain with an occluded peptide binding site.
Nat.Struct.Biol., 5, 1998
|
|
1KSR
 
 | | THE REPEATING SEGMENTS OF THE F-ACTIN CROSS-LINKING GELATION FACTOR (ABP-120) HAVE AN IMMUNOGLOBULIN FOLD, NMR, 20 STRUCTURES | | Descriptor: | GELATION FACTOR | | Authors: | Fucini, P, Renner, C, Herberhold, C, Noegel, A.A, Holak, T.A. | | Deposit date: | 1997-02-07 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The repeating segments of the F-actin cross-linking gelation factor (ABP-120) have an immunoglobulin-like fold.
Nat.Struct.Biol., 4, 1997
|
|
1SVQ
 
 | |
1S0P
 
 | | Structure of the N-Terminal Domain of the Adenylyl Cyclase-Associated Protein (CAP) from Dictyostelium discoideum. | | Descriptor: | Adenylyl cyclase-associated protein, MAGNESIUM ION | | Authors: | Ksiazek, D, Brandstetter, H, Israel, L, Bourenkov, G.P, Katchalova, G, Janssen, K.P, Bartunik, H.D, Noegel, A.A, Schleicher, M, Holak, T.A. | | Deposit date: | 2004-01-01 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | STRUCTURE OF THE N-TERMINAL DOMAIN OF THE ADENYLYL
CYCLASE-ASSOCIATED PROTEIN (CAP) FROM DICTYOSTELIUM DISCOIDEUM
Structure, 11, 2003
|
|
