3HIV
 
 | | Crystal structure of Saporin-L1 in complex with the trinucleotide inhibitor, a transition state analogue | | Descriptor: | (2R,3R,4R,5R)-5-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)-2-({[(S)-({(3R,4R)-4-({[(S)-{[(2R,3R,4R,5R)-5-(2-amino-6-oxo-6,8-dihydro-9H-purin-9-yl)-2-(hydroxymethyl)-4-methoxytetrahydrofuran-3-yl]oxy}(hydroxy)phosphoryl]oxy}methyl)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]pyrrolidin-3-yl}oxy)(hydroxy)phosphoryl]oxy}methyl)-4-methoxytetrahydrofuran-3-yl 3-hydroxypropyl hydrogen (S)-phosphate, Vacuolar saporin | | Authors: | Ho, M, Sturm, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HIT
 
 | | Crystal structure of Saporin-L1 in complex with the dinucleotide inhibitor, a transition state analogue | | Descriptor: | 5'-O-[(S)-{[(3R,4R)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-({[(S)-hydroxy(3-hydroxypropoxy)phosphoryl]oxy}methyl)pyrrolidin-3-yl]oxy}(hydroxy)phosphoryl]-3'-O-[(R)-hydroxy(4-hydroxybutoxy)phosphoryl]-2'-O-methylguanosine, Vacuolar saporin | | Authors: | Ho, M, Sturm, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5YTA
 
 | |
4G56
 
 | | Crystal Structure of full length PRMT5/MEP50 complexes from Xenopus laevis | | Descriptor: | Hsl7 protein, MGC81050 protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Ho, M, Wilczek, C, Bonanno, J, Shechter, D, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity
Plos One, 8, 2013
|
|
3PHC
 
 | | Crystal Structure of Plasmodium falciparum purine nucleoside phosphorylase in complex with DADMe-ImmG | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Ho, M, Edwards, A.A, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-11-03 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Plasmodium falciparum purine nucleoside phosphorylase in complex with DADMe-ImmG
To be Published
|
|
3HIS
 
 | |
3HIO
 
 | | Crystal structure of Ricin A-chain in complex with the cyclic tetranucleotide inhibitor, a transition state analogue | | Descriptor: | 9,9'-{(2R,3R,3aR,5S,7aR,9R,10R,10aR,12S,23R,25aR,27R,28R,28aR,30S,32aR,35aR,37S,39aR)-9-(6-amino-9H-purin-9-yl)-34-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-5,12,23,30,37-pentahydroxy-3,10,28-trimethoxy-5,12,23,30,37-pentaoxidotetracosahydro-2H,7H,25H-trifuro[3,2-f:3',2'-l:3'',2''-x]pyrrolo[3,4-r][1,3,5,9,11,15,17,21,23,27,29,2,4,10,16,22,28]undecaoxazapentaphosphacyclopentatriacontine-2,27-diyl}bis(2-amino-3,9-dihydro-6H-purin-6-one), Ricin, SULFATE ION | | Authors: | Ho, M, Sturm, M.B, Goldman, J.D, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HIW
 
 | | Crystal structure of Saporin-L1 in complex with the cyclic tetranucleotide inhibitor, a transition state analogue | | Descriptor: | 9,9'-{(2R,3R,3aR,5S,7aR,9R,10R,10aR,12S,23R,25aR,27R,28R,28aR,30S,32aR,35aR,37S,39aR)-9-(6-amino-9H-purin-9-yl)-34-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-5,12,23,30,37-pentahydroxy-3,10,28-trimethoxy-5,12,23,30,37-pentaoxidotetracosahydro-2H,7H,25H-trifuro[3,2-f:3',2'-l:3'',2''-x]pyrrolo[3,4-r][1,3,5,9,11,15,17,21,23,27,29,2,4,10,16,22,28]undecaoxazapentaphosphacyclopentatriacontine-2,27-diyl}bis(2-amino-3,9-dihydro-6H-purin-6-one), Vacuolar saporin | | Authors: | Ho, M, Sturm, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3D0O
 
 | |
4EB8
 
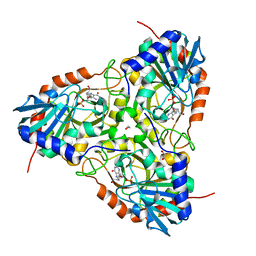 | | Crystal structure of purine nucleoside phosphorylase (W16Y, W94Y, W178Y, H257W) mutant from human complexed with DADMe-ImmG and phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, ... | | Authors: | Ho, M.C, Haapalainen, A.M, Suarez, J.J, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-03-23 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic Site Conformations in Human PNP by (19)F-NMR and Crystallography.
Chem.Biol., 20, 2013
|
|
3EEI
 
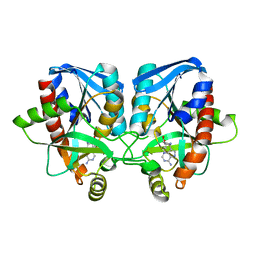 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from neisseria meningitidis in complex with methylthio-immucillin-A | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, 5-methylthioadenosine nucleosidase/S-adenosylhomocysteine nucleosidase | | Authors: | Ho, M, Rinaldo-matthis, A, Brown, R.L, Norris, G.E, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2008-09-04 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from neisseria meningitidis in complex with methylthio-immucillin-A
To be Published
|
|
7WUN
 
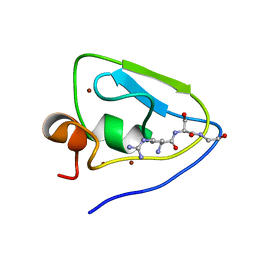 | |
7WUK
 
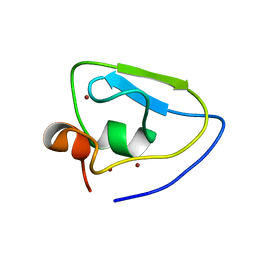 | | Crystal structure of UBR bof from PRT6 | | Descriptor: | E3 ubiquitin-protein ligase, ZINC ION | | Authors: | Ho, M.C, Lin, T.J. | | Deposit date: | 2022-02-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of UBR box from PRT6
To Be Published
|
|
7WUM
 
 | |
7WUL
 
 | |
3U96
 
 | | Crystal Structure of YopHQ357F(Catalytic Domain, Residues 163-468) in complex with pNCS | | Descriptor: | N,4-DIHYDROXY-N-OXO-3-(SULFOOXY)BENZENAMINIUM, SULFATE ION, Tyrosine-protein phosphatase yopH | | Authors: | Ho, M.C, Ke, S. | | Deposit date: | 2011-10-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigation of catalytic loop structure, dynamics, and function relationship of Yersinia protein tyrosine phosphatase by temperature-jump relaxation spectroscopy and X-ray structural determination.
J.Phys.Chem.B, 116, 2012
|
|
4MQ9
 
 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme in complex with GE23077 | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ho, M.X, Arnold, E, Ebright, R.H, Zhang, Y, Tuske, S. | | Deposit date: | 2013-09-16 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
3H3J
 
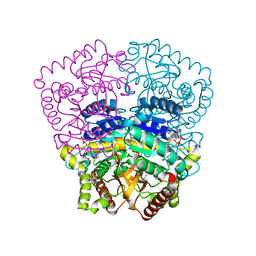 | |
5WSL
 
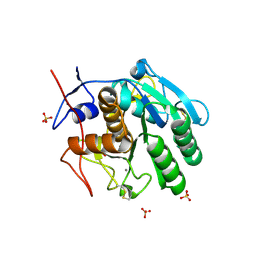 | | Structural studies of keratinase from Meiothermus taiwanensis WR-220 | | Descriptor: | CALCIUM ION, SULFATE ION, keratinase | | Authors: | Ho, M.C, Wu, S.H, Chen, M.Y, Tu, I.F. | | Deposit date: | 2016-12-07 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The discovery of novel heat-stable keratinases from Meiothermus taiwanensis WR-220 and other extremophiles
Sci Rep, 7, 2017
|
|
3CHG
 
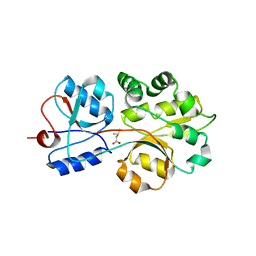 | | The compatible solute-binding protein OpuAC from Bacillus subtilis in complex with DMSA | | Descriptor: | (dimethyl-lambda~4~-sulfanyl)acetic acid, Glycine betaine-binding protein | | Authors: | Smits, S.H.J, Hoing, M, Lecher, J, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2008-03-09 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Compatible-Solute-Binding Protein OpuAC from Bacillus subtilis: Ligand Binding, Site-Directed Mutagenesis, and Crystallographic Studies
J.Bacteriol., 190, 2008
|
|
7T62
 
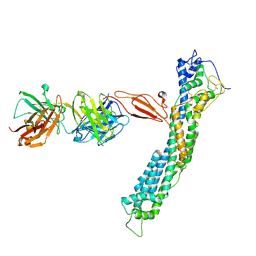 | | GPC2 HEP CT3 complex | | Descriptor: | CT3, Glypican-2 | | Authors: | Zhu, J, Cachau, R, De Val Alda, N, Li, N, Ho, M. | | Deposit date: | 2021-12-13 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | CAR T cells targeting tumor-associated exons of glypican 2 regress neuroblastoma in mice.
Cell Rep Med, 2, 2021
|
|
8IMI
 
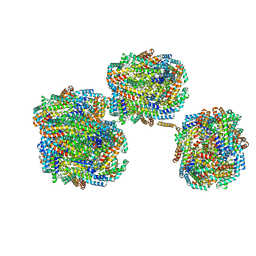 | | A1-A2, A3-A4, B'1-B'2, C'1-C'2 cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster A) | | Descriptor: | ApcA2, ApcB2, ApcB3, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMK
 
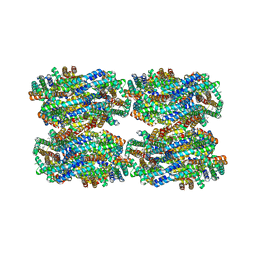 | | D3-D4, D1-D2, D'3-D'4, D'1-D'2 cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster C) | | Descriptor: | ApcA1, ApcA2, ApcB1, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IML
 
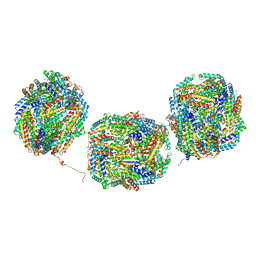 | | Rs2I-Rs2II, Rs1I-Rs1II, RbI-RbII cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster D) | | Descriptor: | CpcA, CpcB, CpcD, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMJ
 
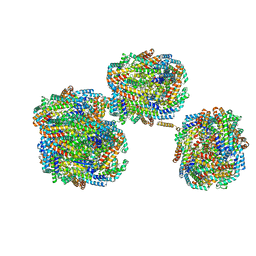 | | A'1-A'2, A'3-A'4, B1-B2, C1-C2 cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster B) | | Descriptor: | ApcA2, ApcB2, ApcB3, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
