4BHW
 
 | | Structural basis for autoinhibition of the acetyltransferase activity of p300 | | Descriptor: | HISTONE ACETYLTRANSFERASE P300, ZINC ION, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate | | Authors: | Delvecchio, M, Gaucher, J, Aguilar-Gurrieri, C, Ortega, E, Panne, D. | | Deposit date: | 2013-04-08 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structure of the P300 Catalytic Core and Implications for Chromatin Targeting and Hat Regulation
Nat.Struct.Mol.Biol., 20, 2013
|
|
5XCZ
 
 | | Structure of the cellobiohydrolase Cel6A from Phanerochaete chrysosporium in complex with cellobiose at 2.1 angstrom | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucanase, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Tachioka, M, Nakamura, A, Ishida, T, Igarashi, K, Samejima, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-07-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a family 6 cellobiohydrolase from the basidiomycete Phanerochaete chrysosporium
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5XCY
 
 | | Structure of the cellobiohydrolase Cel6A from Phanerochaete chrysosporium at 1.2 angstrom | | Descriptor: | Glucanase | | Authors: | Tachioka, M, Nakamura, A, Ishida, T, Igarashi, K, Samejima, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Crystal structure of a family 6 cellobiohydrolase from the basidiomycete Phanerochaete chrysosporium
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3A4U
 
 | | Crystal structure of MCFD2 in complex with carbohydrate recognition domain of ERGIC-53 | | Descriptor: | CALCIUM ION, GLYCEROL, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Nishio, M, Kamiya, Y, Mizushima, T, Wakatsuki, S, Sasakawa, H, Yamamoto, K, Uchiyama, S, Noda, M, McKay, A.R, Fukui, K, Hauri, H.P, Kato, K. | | Deposit date: | 2009-07-17 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the cooperative interplay between the two causative gene products of combined factor V and factor VIII deficiency.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2LTP
 
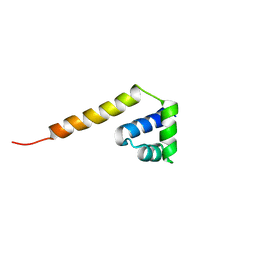 | | Solution structure of the SANT2 domain of the human nuclear receptor corepressor 2 (NCoR2), Northeast Structural Genomics Consortium (NESG) target ID HR4636E | | Descriptor: | Nuclear receptor corepressor 2 | | Authors: | Montecchio, M, Lemak, A, Yee, A, Xu, C, Garcia, M, Houliston, S, Bellanda, M, Min, J, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SANT2 domain of the human nuclear receptor corepressor 2 (NCoR2).
To be Published
|
|
2LLK
 
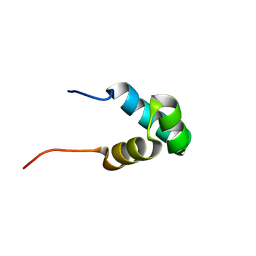 | | Solution NMR structure of the N-terminal myb-like 1 domain of the human cyclin-D-binding transcription factor 1 (hDMP1), Northeast Structural Genomics Consortium (NESG) target ID hr8011a | | Descriptor: | Cyclin-D-binding Myb-like transcription factor 1 | | Authors: | Montecchio, M, Lemak, A, Yee, A, Xu, C, Garcia, M, Houliston, S, Min, J, Bellanda, M, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-10 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the N-terminal myb-like 1 domain of the human cyclin D binding transcription factor 1 (hDMP1).
To be Published
|
|
7VEW
 
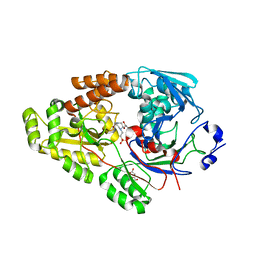 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with unsaturated trigalacturonic acid | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEQ
 
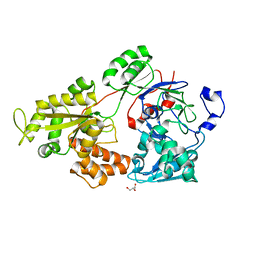 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in an open conformation | | Descriptor: | GLYCEROL, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEV
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VER
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a full open conformation | | Descriptor: | GLYCEROL, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VET
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a closed conformation | | Descriptor: | SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEU
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with galacturonic acid | | Descriptor: | GLYCEROL, SPH1118, alpha-D-galactopyranuronic acid | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
4KMI
 
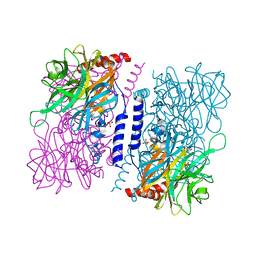 | | Crystal structure of 4-O-beta-D-mannosyl-D-glucose phosphorylase MGP complexed with PO4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-O-beta-D-mannosyl-D-glucose phosphorylase, PHOSPHATE ION | | Authors: | Nakae, S, Ito, S, Higa, M, Senoura, T, Wasaki, J, Hijikata, A, Shionyu, M, Ito, S, Shirai, T. | | Deposit date: | 2013-05-08 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Novel Enzyme in Mannan Biodegradation Process 4-O-beta-d-Mannosyl-d-Glucose Phosphorylase MGP
J.Mol.Biol., 425, 2013
|
|
2YR2
 
 | |
8TO0
 
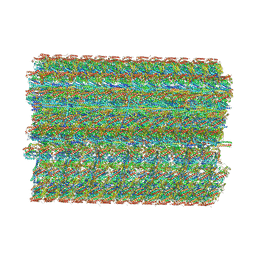 | | 48-nm repeating structure of doublets from mouse sperm flagella | | Descriptor: | Cilia- and flagella- associated protein 210, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Chen, Z, Shiozak, M, Hass, K.M, Skinner, W, Zhao, S, Guo, C, Polacco, B.J, Yu, Z, Krogan, N.J, Kaake, R.M, Vale, R.D, Agard, D.A. | | Deposit date: | 2023-08-02 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | De novo protein identification in mammalian sperm using in situ cryoelectron tomography and AlphaFold2 docking.
Cell, 186, 2023
|
|
7QBR
 
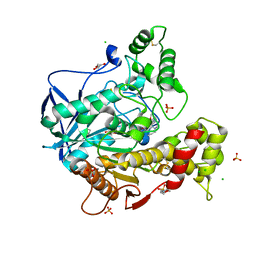 | | Human butyrylcholinesterase in complex with (Z)-N-tert-butyl-1-(8-(3-(4-(prop-2-yn-1-yl)piperazin-1-yl)propoxy)quinolin-2-yl)methanimine oxide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Denic, M, Chioua, M, Knez, D, Gobec, S, Nachon, F, Marco-Contelles, J.L, Brazzolotto, X. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | 8-Hydroxyquinolylnitrones as multifunctional ligands for the therapy of neurodegenerative diseases.
Acta Pharm Sin B, 13, 2023
|
|
7QBQ
 
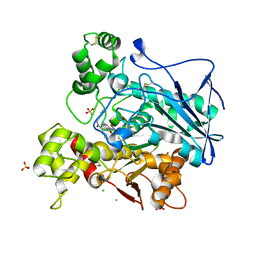 | | Human butyrylcholinesterase in complex with (Z)-N-benzyl-1-(8-hydroxyquinolin-2-yl)methanimine oxide | | Descriptor: | 1-(8-oxidanylquinolin-2-yl)-N-(phenylmethyl)methanimine oxide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Denic, M, Chioua, M, Knez, D, Gobec, S, Nachon, F, Marco-Contelles, J.L, Brazzolotto, X. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | 8-Hydroxyquinolylnitrones as multifunctional ligands for the therapy of neurodegenerative diseases.
Acta Pharm Sin B, 13, 2023
|
|
5YRM
 
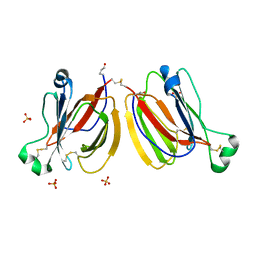 | | PPL3C-isomaltose complex | | Descriptor: | PPL3-b, SULFATE ION, alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose | | Authors: | Nakae, S, Shionyu, M, Ogawa, T, Shirai, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization
Proteins, 86, 2018
|
|
5YRE
 
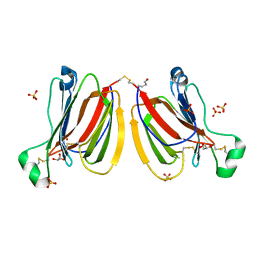 | | Crystal structure of PPL3A | | Descriptor: | GLYCEROL, PPL3-A, SULFATE ION | | Authors: | Nakae, S, Shionyu, M, Ogawa, T, Shirai, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization
Proteins, 86, 2018
|
|
4W2I
 
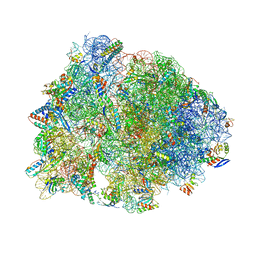 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with negamycin, mRNA and three deacylated tRNAs in the A, P and E sites | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Szal, T, Jiang, F, Gupta, P, Matsuda, R, Shiozuka, M, Steitz, T.A, Vazquez-Laslop, N, Mankin, A.S. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Negamycin Interferes with Decoding and Translocation by Simultaneous Interaction with rRNA and tRNA.
Mol.Cell, 56, 2014
|
|
7NFW
 
 | |
7NIZ
 
 | | Human 14-3-3 sigma in complex with human Estrogen Receptor alpha peptide and ligands Fusicoccin-A and WR-1065 | | Descriptor: | 14-3-3 protein sigma, Estrogen receptor, FUSICOCCIN, ... | | Authors: | Roversi, P, Falcicchio, M, Doveston, R. | | Deposit date: | 2021-02-14 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Cooperative stabilisation of 14-3-3 sigma protein-protein interactions via covalent protein modification.
Chem Sci, 12, 2021
|
|
5YRK
 
 | | Crystal structure of PPL3C | | Descriptor: | PPL3-b, SULFATE ION | | Authors: | Nakae, S, Shionyu, M, Ogawa, T, Shirai, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-08-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization
Proteins, 86, 2018
|
|
5YRH
 
 | | Crystal structure of PPL3B | | Descriptor: | PPL3-a, PPL3-b, SULFATE ION | | Authors: | Nakae, S, Shionyu, M, Ogawa, T, Shirai, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization
Proteins, 86, 2018
|
|
5YRI
 
 | | PPL3B-trehalose complex | | Descriptor: | PPL3-a, PPL3-b, SULFATE ION, ... | | Authors: | Nakae, S, Shionyu, M, Ogawa, T, Shirai, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization
Proteins, 86, 2018
|
|
