7OF6
 
 | | Structure of mature human mitochondrial ribosome large subunit in complex with GTPBP6 (PTC conformation 2). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
7OF0
 
 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with MTERF4-NSUN4 (dataset1). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
7OF2
 
 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with GTPBP6. | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
7OF4
 
 | | Structure of mature human mitochondrial ribosome large subunit in complex with GTPBP6 (PTC conformation 1). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
7OF5
 
 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with MTERF4-NSUN4 and GTPBP5 (dataset2). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
7OF7
 
 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with MTERF4-NSUN4 and GTPBP5 (dataset1). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
5OL9
 
 | | Structure of human mitochondrial transcription elongation factor (TEFM) N-terminal domain | | Descriptor: | ACETATE ION, Transcription elongation factor, mitochondrial | | Authors: | Hillen, H.S, Parshin, A.V, Agaronyan, K, Morozov, Y, Graber, J.J, Chernev, A, Schwinghammer, K, Urlaub, H, Anikin, M, Cramer, P, Temiakov, D. | | Deposit date: | 2017-07-27 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Mechanism of Transcription Anti-termination in Human Mitochondria.
Cell, 171, 2017
|
|
5OL8
 
 | | Structure of human mitochondrial transcription elongation factor (TEFM) C-terminal domain | | Descriptor: | GLYCEROL, Transcription elongation factor, mitochondrial | | Authors: | Hillen, H.S, Parshin, A.V, Agaronyan, K, Morozov, Y, Graber, J.J, Chernev, A, Schwinghammer, K, Urlaub, H, Anikin, M, Cramer, P, Temiakov, D. | | Deposit date: | 2017-07-27 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Transcription Anti-termination in Human Mitochondria.
Cell, 171, 2017
|
|
5OLA
 
 | | Structure of mitochondrial transcription elongation complex in complex with elongation factor TEFM | | Descriptor: | DNA (30-MER), DNA (5'-D(P*AP*TP*GP*GP*TP*GP*TP*AP*AP*CP*GP*CP*CP*AP*GP*AP*CP*GP*AP*AP*C)-3'), DNA-directed RNA polymerase, ... | | Authors: | Hillen, H.S, Parshin, A.V, Agaronyan, K, Morozov, Y, Graber, J.J, Chernev, A, Schwinghammer, K, Urlaub, H, Anikin, M, Cramer, P, Temiakov, D. | | Deposit date: | 2017-07-27 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.904 Å) | | Cite: | Mechanism of Transcription Anti-termination in Human Mitochondria.
Cell, 171, 2017
|
|
7OF3
 
 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with MTERF4-NSUN4 (dataset2). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
6YYT
 
 | | Structure of replicating SARS-CoV-2 polymerase | | Descriptor: | RNA product, ZINC ION, nsp12, ... | | Authors: | Hillen, H.S, Kokic, G, Farnung, L, Dienemann, C, Tegunov, D, Cramer, P. | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of replicating SARS-CoV-2 polymerase.
Nature, 584, 2020
|
|
7A8P
 
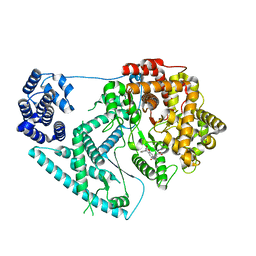 | | Structure of human mitochondrial RNA polymerase in complex with IMT inhibitor. | | Descriptor: | (3~{R})-1-[(2~{R})-2-[4-(2-chloranyl-4-fluoranyl-phenyl)-2-oxidanylidene-chromen-7-yl]oxypropanoyl]piperidine-3-carboxylic acid, DNA-directed RNA polymerase, mitochondrial | | Authors: | Hillen, H.S, Bonekamp, N, Peter, B, Felser, A, Bergbrede, T, Choidas, A, Horn, M, Unger, A, di Lucrezia, R, Atanassov, I, Li, X, Koch, U, Menninger, S, Boros, J, Habenberger, P, Giavalisco, P, Cramer, P, Denzel, M, Nussbaumer, P, Klebl, B, Falkenberg, M, Gustafsson, C.M, Larsson, N.G. | | Deposit date: | 2020-08-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Small-molecule inhibitors of human mitochondrial DNA transcription.
Nature, 588, 2020
|
|
7A9W
 
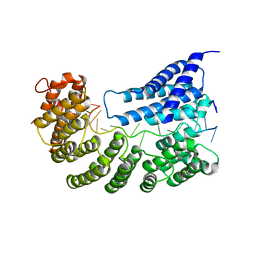 | | Structure of yeast Rmd9p in complex with 20nt target RNA | | Descriptor: | CHLORIDE ION, Protein RMD9, mitochondrial, ... | | Authors: | Hillen, H.S, Markov, D.A, Ireneusz, W.D, Hofmann, K.B, Cowan, A.T, Jones, J.L, Temiakov, D, Cramer, P, Anikin, M. | | Deposit date: | 2020-09-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The pentatricopeptide repeat protein Rmd9 recognizes the dodecameric element in the 3'-UTRs of yeast mitochondrial mRNAs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7A9X
 
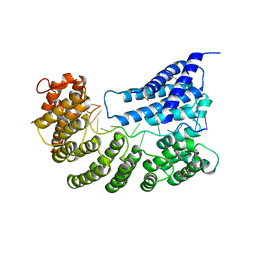 | | Structure of yeast Rmd9p in complex with 16nt target RNA | | Descriptor: | CHLORIDE ION, Protein RMD9, mitochondrial, ... | | Authors: | Hillen, H.S, Markov, D.A, Ireneusz, W.D, Hofmann, K.B, Cowan, A.T, Jones, J.L, Temiakov, D, Cramer, P, Anikin, M. | | Deposit date: | 2020-09-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The pentatricopeptide repeat protein Rmd9 recognizes the dodecameric element in the 3'-UTRs of yeast mitochondrial mRNAs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ERQ
 
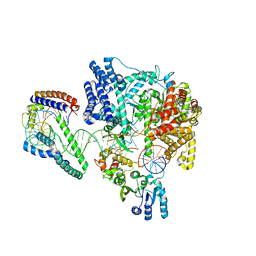 | | Structure of the human mitochondrial transcription initiation complex at the HSP promoter | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, Dimethyladenosine transferase 2, ... | | Authors: | Hillen, H.S, Morozov, Y.I, Sarfallah, A, Temiakov, D, Cramer, P. | | Deposit date: | 2017-10-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural Basis of Mitochondrial Transcription Initiation.
Cell, 171, 2017
|
|
6RIE
 
 | | Structure of Vaccinia Virus DNA-dependent RNA polymerase co-transcriptional capping complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Hillen, H.S, Bartuli, J, Grimm, C, Dienemann, C, Bedenk, K, Szalar, A, Fischer, U, Cramer, P. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
6RID
 
 | | Structure of Vaccinia Virus DNA-dependent RNA polymerase elongation complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Hillen, H.S, Bartuli, J, Grimm, C, Dienemann, C, Bedenk, K, Szalar, A, Fischer, U, Cramer, P. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
6ERP
 
 | | Structure of the human mitochondrial transcription initiation complex at the LSP promoter | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, Dimethyladenosine transferase 2, ... | | Authors: | Hillen, H.S, Morozov, Y.I, Sarfallah, A, Temiakov, D, Cramer, P. | | Deposit date: | 2017-10-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.502 Å) | | Cite: | Structural Basis of Mitochondrial Transcription Initiation.
Cell, 171, 2017
|
|
6ERO
 
 | | Structure of human TFB2M | | Descriptor: | CHLORIDE ION, Dimethyladenosine transferase 2, mitochondrial,Dimethyladenosine transferase 2, ... | | Authors: | Hillen, H.S, Morozov, Y.I, Sarfallah, A, Temiakov, D, Cramer, P. | | Deposit date: | 2017-10-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Mitochondrial Transcription Initiation.
Cell, 171, 2017
|
|
9GJU
 
 | | Structure of replicating Nipah Virus RNA Polymerase Complex - RNA-bound state | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Phosphoprotein, ... | | Authors: | Sala, F, Ditter, K, Dybkov, O, Urlaub, H, Hillen, H.S. | | Deposit date: | 2024-08-22 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of Nipah virus RNA synthesis.
Nat Commun, 16, 2025
|
|
9GJT
 
 | | Structure of Nipah Virus RNA Polymerase Complex - Apo state | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Sala, F, Ditter, K, Dybkov, O, Urlaub, H, Hillen, H.S. | | Deposit date: | 2024-08-22 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of Nipah virus RNA synthesis.
Nat Commun, 16, 2025
|
|
6RIC
 
 | | Structure of the core Vaccinia Virus DNA-dependent RNA polymerase complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Grimm, C, Hillen, H.S, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A, Cramer, P, Fischer, U. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
1MD7
 
 | | Monomeric structure of the zymogen of complement protease C1r | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C1R COMPLEMENT SERINE PROTEASE | | Authors: | Budayova-Spano, M, Grabarse, W, Thielens, N.M, Hillen, H, Lacroix, M, Schmidt, M, Fontecilla-Camps, J, Arlaud, G.J, Gaboriaud, C. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Monomeric structures of the zymogen and active catalytic domain of complement protease c1r: further insights into the c1 activation mechanism
Structure, 10, 2002
|
|
1MD8
 
 | | Monomeric structure of the active catalytic domain of complement protease C1r | | Descriptor: | C1R COMPLEMENT SERINE PROTEASE | | Authors: | Budayova-Spano, M, Grabarse, W, Thielens, N.M, Hillen, H, Lacroix, M, Schmidt, M, Fontecilla-Camps, J, Arlaud, G.J, Gaboriaud, C. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Monomeric structures of the zymogen and active catalytic domain of complement protease c1r: further insights into the c1 activation mechanism
Structure, 10, 2002
|
|
8QMA
 
 | | Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | do Prado, P.F.V, Ahrens, F.M, Pfannschmidt, T, Hillen, H.S. | | Deposit date: | 2023-09-21 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the multi-subunit chloroplast RNA polymerase.
Mol.Cell, 84, 2024
|
|
