5ACL
 
 | | Mcg immunoglobulin variable domain with sulfasalazine | | Descriptor: | 2-HYDROXY-(5-([4-(2-PYRIDINYLAMINO)SULFONYL]PHENYL)AZO)BENZOIC ACID, MCG, SULFATE ION | | Authors: | Brumshtein, B, Esswein, S.R, Salwinski, L, Phillips, M.L, Ly, A.T, Cascio, D, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Inhibition by small-molecule ligands of formation of amyloid fibrils of an immunoglobulin light chain variable domain.
Elife, 4, 2015
|
|
6DFT
 
 | | Trypanosoma brucei deoxyhypusine synthase | | Descriptor: | Deoxyhypusine synthase, Deoxyhypusine synthase regulatory subunit, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Tomchick, D.R, Phillips, M.A, Afanador, G.A. | | Deposit date: | 2018-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Trypanosomatid Deoxyhypusine Synthase Activity Is Dependent on Shared Active-Site Complementation between Pseudoenzyme Paralogs.
Structure, 26, 2018
|
|
3FGH
 
 | | Human mitochondrial transcription factor A box B | | Descriptor: | CADMIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Gangelhoff, T.A, Mungalachetty, P, Nix, J, Churchill, M.E.A. | | Deposit date: | 2008-12-06 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A
Nucleic Acids Res., 37, 2009
|
|
6E0B
 
 | | Plasmodium falciparum dihydroorotate dehydrogenase C276F mutant bound with triazolopyrimidine-based inhibitor DSM1 | | Descriptor: | 5-methyl-7-(naphthalen-2-ylamino)-1H-[1,2,4]triazolo[1,5-a]pyrimidine-3,8-diium, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Tomchick, D.R, Phillips, M.A, Deng, X. | | Deposit date: | 2018-07-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Identification and Mechanistic Understanding of Dihydroorotate Dehydrogenase Point Mutations in Plasmodium falciparum that Confer in Vitro Resistance to the Clinical Candidate DSM265.
ACS Infect Dis, 5, 2019
|
|
5ACM
 
 | | Mcg immunoglobulin variable domain with methylene blue | | Descriptor: | 3,7-BIS(DIMETHYLAMINO)PHENOTHIAZIN-5-IUM, GLYCEROL, MCG, ... | | Authors: | Brumshtein, B, Esswein, S.R, Salwinski, L, Phillips, M.L, Ly, A.T, Cascio, D, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Inhibition by small-molecule ligands of formation of amyloid fibrils of an immunoglobulin light chain variable domain.
Elife, 4, 2015
|
|
5OF1
 
 | | The structural versatility of TasA in B. subtilis biofilm formation | | Descriptor: | 1,2-ETHANEDIOL, 2-HYDROXYBENZOIC ACID, Spore coat-associated protein N | | Authors: | Roske, Y, Diehl, A, Ball, L, Chowdhury, A, Hiller, M, Moliere, N, Kramer, R, Nagaraj, M, Stoeppler, D, Worth, C.L, Schlegel, B, Leidert, M, Cremer, N, Eisenmenger, F, Lopez, D, Schmieder, P, Heinemann, U, Turgay, K, Akbey, U, Oschkinat, H. | | Deposit date: | 2017-07-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural changes of TasA in biofilm formation ofBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5OF2
 
 | | The structural versatility of TasA in B. subtilis biofilm formation | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Spore coat-associated protein N | | Authors: | Roske, Y, Diehl, A, Ball, L, Chowdhury, A, Hiller, M, Moliere, N, Kramer, R, Nagaraj, M, Stoeppler, D, Worth, C.L, Schlegel, B, Leidert, M, Cremer, N, Eisenmenger, F, Lopez, D, Schmieder, P, Heinemann, U, Turgay, K, Akbey, U, Oschkinat, H. | | Deposit date: | 2017-07-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural changes of TasA in biofilm formation ofBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3I65
 
 | | Plasmodium falciparum dihydroorotate dehydrogenase bound with triazolopyrimidine-based inhibitor DSM1 | | Descriptor: | 5-methyl-7-(naphthalen-2-ylamino)-1H-[1,2,4]triazolo[1,5-a]pyrimidine-3,8-diium, Dihydroorotate dehydrogenase homolog, mitochondrial, ... | | Authors: | Deng, X, Phillips, M.A. | | Deposit date: | 2009-07-06 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural plasticity of malaria dihydroorotate dehydrogenase allows selective binding of diverse chemical scaffolds.
J.Biol.Chem., 284, 2009
|
|
1HMA
 
 | | THE SOLUTION STRUCTURE AND DYNAMICS OF THE DNA BINDING DOMAIN OF HMG-D FROM DROSOPHILA MELANOGASTER | | Descriptor: | HMG-D | | Authors: | Jones, D.N.M, Searles, M.A, Shaw, G.L, Churchill, M.E.A, Ner, S.S, Keeler, J, Travers, A.A, Neuhaus, D. | | Deposit date: | 1994-05-12 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of the DNA-binding domain of HMG-D from Drosophila melanogaster.
Structure, 2, 1994
|
|
3ZEV
 
 | | Structure of Thermostable Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | GLYCINE, NEUROTENSIN, NEUROTENSIN RECEPTOR 1 TM86V | | Authors: | Egloff, P, Hillenbrand, M, Schlinkmann, K.M, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2012-12-07 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3I68
 
 | |
3I6R
 
 | | Plasmodium falciparum dihydroorotate dehydrogenase bound with triazolopyrimidine-based inhibitor DSM74 | | Descriptor: | 5-methyl-N-[4-(trifluoromethyl)phenyl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase homolog, mitochondrial, ... | | Authors: | Deng, X, Phillips, M.A. | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural plasticity of malaria dihydroorotate dehydrogenase allows selective binding of diverse chemical scaffolds.
J.Biol.Chem., 284, 2009
|
|
1F3T
 
 | | CRYSTAL STRUCTURE OF TRYPANOSOMA BRUCEI ORNITHINE DECARBOXYLASE (ODC) COMPLEXED WITH PUTRESCINE, ODC'S REACTION PRODUCT. | | Descriptor: | 1,4-DIAMINOBUTANE, ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jackson, L.K, Brooks, H.B, Osterman, A.L, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2000-06-06 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Altering the reaction specificity of eukaryotic ornithine decarboxylase.
Biochemistry, 39, 2000
|
|
2C7A
 
 | | STRUCTURE OF THE PROGESTERONE RECEPTOR-DNA COMPLEX | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*AP*AP *CP*TP*GP*TP*TP*CP*TP*G)-3', 5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*TP *TP*TP*GP*TP*TP*CP*TP*G)-3', PROGESTERONE RECEPTOR, ... | | Authors: | Roemer, S.C, Donham, D.C, Sherman, L, Pon, V.H, Edwards, D.P, Churchill, M.E.A. | | Deposit date: | 2005-11-19 | | Release date: | 2006-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Progesterone Receptor-Deoxyribonucleic Acid Complex: Novel Interactions Required for Binding to Half-Site Response Elements.
Mol.Endocrinol., 20, 2006
|
|
3O8A
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with novel Inhibitor Genz667348 | | Descriptor: | Dihydroorotate dehydrogenase homolog, mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Booker, M.L, Phillips, M.A. | | Deposit date: | 2010-08-02 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel inhibitors of Plasmodium falciparum dihydroorotate dehydrogenase with anti-malarial activity in the mouse model.
J.Biol.Chem., 285, 2010
|
|
1F0M
 
 | | MONOMERIC STRUCTURE OF THE HUMAN EPHB2 SAM (STERILE ALPHA MOTIF) DOMAIN | | Descriptor: | EPHRIN TYPE-B RECEPTOR 2 | | Authors: | Thanos, C.D, Faham, S, Goodwill, K.E, Cascio, D, Phillips, M, Bowie, J.U. | | Deposit date: | 2000-05-16 | | Release date: | 2000-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Monomeric structure of the human EphB2 sterile alpha motif domain.
J.Biol.Chem., 274, 1999
|
|
2X9Y
 
 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
2A18
 
 | | carboxysome shell protein ccmK4, crystal form 2 | | Descriptor: | AMMONIUM ION, Carbon dioxide concentrating mechanism protein ccmK homolog 4 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Tanaka, S, Nguyen, C.V, Phillips, M, Beeby, M, Yeates, T.O. | | Deposit date: | 2005-06-18 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Protein structures forming the shell of primitive bacterial organelles
Science, 309, 2005
|
|
2X9X
 
 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, IMIDAZOLE, SODIUM ION | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
2X9Z
 
 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
3N2O
 
 | | X-ray crystal structure of arginine decarboxylase complexed with Arginine from Vibrio vulnificus | | Descriptor: | AGMATINE, Biosynthetic arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-18 | | Release date: | 2010-06-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
2X9W
 
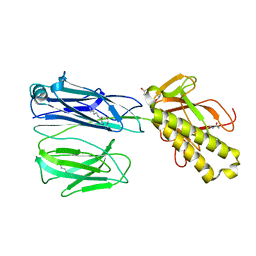 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, GLYCEROL | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
2A1B
 
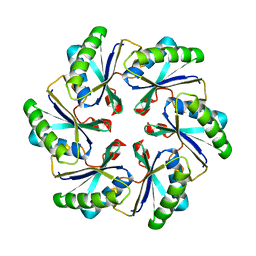 | | Carboxysome shell protein ccmK2 | | Descriptor: | Carbon dioxide concentrating mechanism protein ccmK homolog 2 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Tanaka, S, Nguyen, C.V, Phillips, M, Beeby, M, Yeates, T.O. | | Deposit date: | 2005-06-20 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein structures forming the shell of primitive bacterial organelles
Science, 309, 2005
|
|
2A10
 
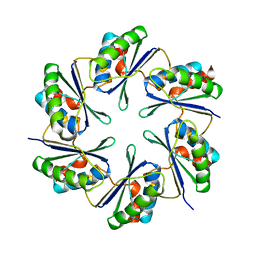 | | carboxysome shell protein ccmK4 | | Descriptor: | Carbon dioxide concentrating mechanism protein ccmK homolog 4 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Tanaka, S, Nguyen, C.V, Phillips, M, Beeby, M, Yeates, T.O. | | Deposit date: | 2005-06-17 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Protein structures forming the shell of primitive bacterial organelles
Science, 309, 2005
|
|
1EG2
 
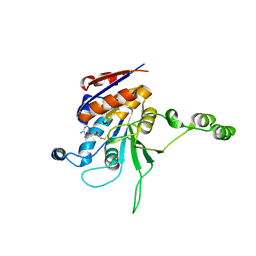 | | CRYSTAL STRUCTURE OF RHODOBACTER SPHEROIDES (N6 ADENOSINE) METHYLTRANSFERASE (M.RSRI) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MODIFICATION METHYLASE RSRI | | Authors: | Scavetta, R.D, Thomas, C.B, Walsh, M.A, Szegedi, S, Joachimiak, A, Gumport, R.I, Churchill, M.E.A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-02-11 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases.
Nucleic Acids Res., 28, 2000
|
|
