6S19
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 4.57 keV from all available diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.Ch. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1D
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 4.57 keV from 20,000 diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1G
 
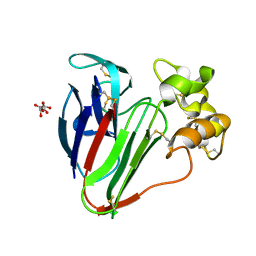 | | Structure of thaumatin determined at SwissFEL using native-SAD at 6.06 keV from 50,000 diffraction patterns. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S0Q
 
 | | Structure of the A2A adenosine receptor determined at SwissFEL using native-SAD at 4.57 keV from 50,000 diffraction patterns | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1E
 
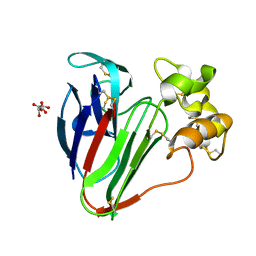 | | Structure of thaumatin determined at SwissFEL using native-SAD at 6.06 keV from all available diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
2M1A
 
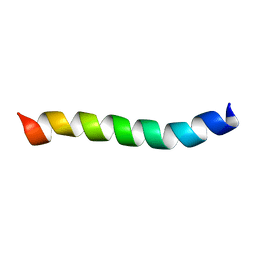 | | HIV-1 Rev ARM peptide (residues T34-R50) | | Descriptor: | HIV-1 Rev arginine-rich motif (ARM) | | Authors: | Casu, F, Duggan, B.M, Hennig, M. | | Deposit date: | 2012-11-21 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Arginine-Rich RNA-Binding Motif of HIV-1 Rev Is Intrinsically Disordered and Folds upon RRE Binding.
Biophys.J., 105, 2013
|
|
2VB9
 
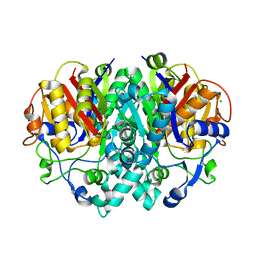 | | beta-ketoacyl-ACP synthase I (KAS) from E. coli, apo structure | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 1, CHLORIDE ION | | Authors: | Pappenberger, G, Schulz-Gasch, T, Bailly, J, Hennig, M. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Assisted Discovery of an Aminothiazole Derivative as a Lead Molecule for Inhibition of Bacterial Fatty-Acid Synthesis.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2VBA
 
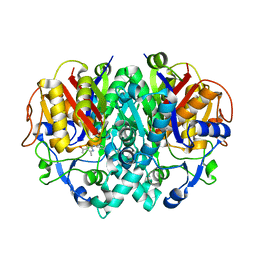 | | beta-ketoacyl-ACP synthase I (KAS) from E. coli with bound amino- thiazole inhibitor | | Descriptor: | 2-PHENYLAMINO-4-METHYL-5-ACETYL THIAZOLE, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 1 | | Authors: | Pappenberger, G, Schulz-Gasch, T, Bailly, J, Hennig, M. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure-Assisted Discovery of an Aminothiazole Derivative as a Lead Molecule for Inhibition of Bacterial Fatty-Acid Synthesis.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2VB7
 
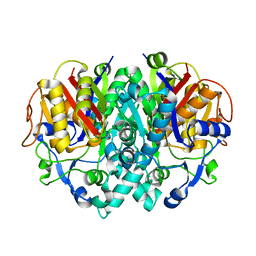 | | beta-ketoacyl-ACP synthase I (KAS) from E. coli, apo structure after soak in PEG solution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 1 | | Authors: | Pappenberger, G, Schulz-Gasch, T, Bailly, J, Hennig, M. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Assisted Discovery of an Aminothiazole Derivative as a Lead Molecule for Inhibition of Bacterial Fatty-Acid Synthesis.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3BUH
 
 | | BACE-1 complexed with compound 4 | | Descriptor: | 4-(2-aminoethyl)-2-cyclohexylphenol, beta-secretase 1 | | Authors: | Kuglstatter, A, Hennig, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-03-11 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tyramine fragment binding to BACE-1
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BRA
 
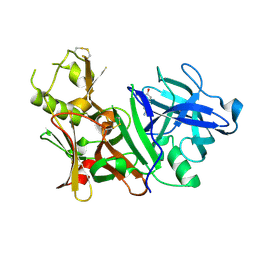 | | BACE-1 complexed with compound 1 | | Descriptor: | 4-(2-aminoethyl)phenol, Beta-secretase 1 | | Authors: | Kuglstatter, A, Hennig, M. | | Deposit date: | 2007-12-21 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tyramine fragment binding to BACE-1
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BUF
 
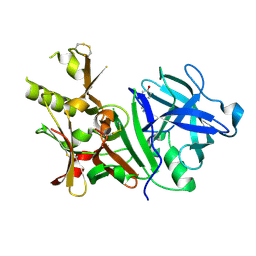 | | BACE-1 complexed with compound 2 | | Descriptor: | 4-[(2R)-2-aminopropyl]phenol, Beta-secretase 1 | | Authors: | Kuglstatter, A, Hennig, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-03-11 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tyramine fragment binding to BACE-1
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BUG
 
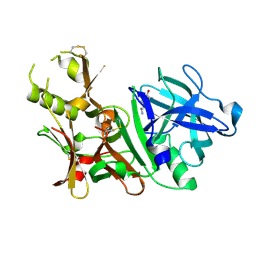 | | BACE-1 complexed with compound 3 | | Descriptor: | 4-(2-aminoethyl)-2-ethylphenol, BETA-SECRETASE 1 | | Authors: | Kuglstatter, A, Hennig, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-03-11 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tyramine fragment binding to BACE-1
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2HVM
 
 | |
2DEB
 
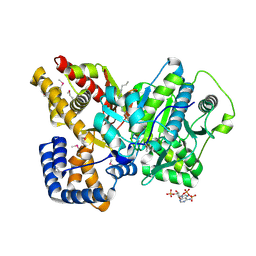 | | Crystal structure of rat carnitine palmitoyltransferase 2 in space group C2221 | | Descriptor: | COENZYME A, Carnitine O-palmitoyltransferase II, mitochondrial, ... | | Authors: | Rufer, A.C, Thoma, R, Benz, J, Stihle, M, Gsell, B, De Roo, E, Banner, D.W, Mueller, F, Chomienne, O, Hennig, M. | | Deposit date: | 2006-02-10 | | Release date: | 2007-02-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of carnitine palmitoyltransferase 2 and implications for diabetes treatment
Structure, 14, 2006
|
|
2FYO
 
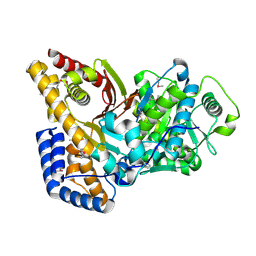 | | Crystal structure of rat carnitine palmitoyltransferase 2 in space group P43212 | | Descriptor: | Carnitine O-palmitoyltransferase II, mitochondrial | | Authors: | Rufer, A.C, Thoma, R, Benz, J, Stihle, M, Gsell, B, De Roo, E, Banner, D.W, Mueller, F, Chomienne, O, Hennig, M. | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-08 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of carnitine palmitoyltransferase 2 and implications for diabetes treatment
Structure, 14, 2006
|
|
1JUK
 
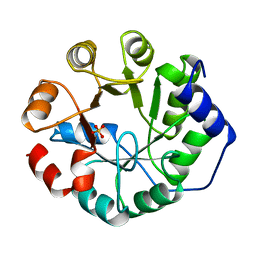 | | INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS IN A TRIGONAL CRYSTAL FORM | | Descriptor: | INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE, SULFATE ION | | Authors: | Knoechel, T.R, Hennig, M, Merz, A, Darimont, B, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1996-05-03 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of indole-3-glycerol phosphate synthase from the hyperthermophilic archaeon Sulfolobus solfataricus in three different crystal forms: effects of ionic strength.
J.Mol.Biol., 262, 1996
|
|
1JUL
 
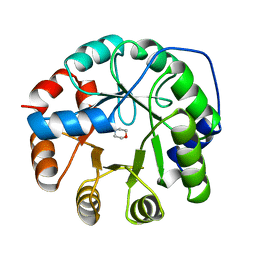 | | INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS IN A SECOND ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | Authors: | Knoechel, T.R, Hennig, M, Merz, A, Darimont, B, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1996-05-03 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of indole-3-glycerol phosphate synthase from the hyperthermophilic archaeon Sulfolobus solfataricus in three different crystal forms: effects of ionic strength.
J.Mol.Biol., 262, 1996
|
|
3S3X
 
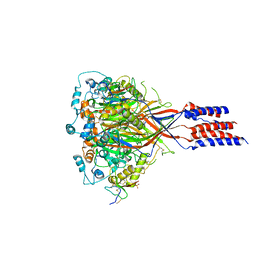 | | Structure of chicken acid-sensing ion channel 1 AT 3.0 A resolution in complex with psalmotoxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive cation channel 2, neuronal, ... | | Authors: | Dawson, R.J.P, Benz, J, Stohler, P, Tetaz, T, Joseph, C, Huber, S, Schmid, G, Huegin, D, Pflimlin, P, Trube, G, Rudolph, M.G, Hennig, M, Ruf, A. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of the Acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1.
Nat Commun, 3, 2012
|
|
3S3W
 
 | | Structure of chicken acid-sensing ion channel 1 at 2.6 a resolution and ph 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive cation channel 2, ... | | Authors: | Dawson, R.J.P, Benz, J, Stohler, P, Tetaz, T, Joseph, C, Huber, S, Schmid, G, Huegin, D, Pflimlin, P, Trube, G, Rudolph, M.G, Hennig, M, Ruf, A. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1.
Nat Commun, 3, 2012
|
|
1CFF
 
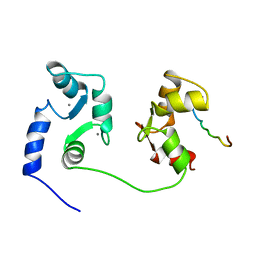 | | NMR SOLUTION STRUCTURE OF A COMPLEX OF CALMODULIN WITH A BINDING PEPTIDE OF THE CA2+-PUMP | | Descriptor: | CALCIUM ION, CALCIUM PUMP, CALMODULIN | | Authors: | Elshorst, B, Hennig, M, Foersterling, H, Diener, A, Maurer, M, Schulte, P, Schwalbe, H, Krebs, J, Schmid, H, Vorherr, T, Carafoli, E, Griesinger, C. | | Deposit date: | 1999-03-18 | | Release date: | 1999-09-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a complex of calmodulin with a binding peptide of the Ca2+ pump.
Biochemistry, 38, 1999
|
|
1DXJ
 
 | | Structure of the chitinase from jack bean | | Descriptor: | CLASS II CHITINASE, SULFATE ION | | Authors: | Hahn, M, Hennig, M, Schlesier, B, Hohne, W. | | Deposit date: | 2000-01-10 | | Release date: | 2000-08-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Jack Bean Chitinase
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2FW3
 
 | | Crystal structure of rat carnitine palmitoyltransferase 2 in complex with antidiabetic drug ST1326 | | Descriptor: | (3R)-3-{[(TETRADECYLAMINO)CARBONYL]AMINO}-4-(TRIMETHYLAMMONIO)BUTANOATE, Carnitine O-palmitoyltransferase II, mitochondrial | | Authors: | Rufer, A.C, Thoma, R, Benz, J, Stihle, M, Gsell, B, De Roo, E, Banner, D.W, Mueller, F, Chomienne, O, Hennig, M. | | Deposit date: | 2006-02-01 | | Release date: | 2007-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of carnitine palmitoyltransferase 2 and implications for diabetes treatment
Structure, 14, 2006
|
|
3SGK
 
 | | Unique carbohydrate/carbohydrate interactions are required for high affinity binding of FcgIII and antibodies lacking core fucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)][2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fc fragment, ... | | Authors: | Ferrara, C, Grau, S, Jaeger, C, Sondermann, P, Bruenker, P, Waldhauer, I, Hennig, M, Ruf, A, Rufer, A.C, Stihle, M, Umana, P, Benz, J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SGJ
 
 | | Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgIII and antibodies lacking core fucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MALONATE ION, ... | | Authors: | Ferrara, C, Grau, S, Jaeger, C, Sondermann, P, Bruenker, P, Waldhauer, I, Hennig, M, Ruf, A, Rufer, A.C, Stihle, M, Umana, P, Benz, J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
