2FZG
 
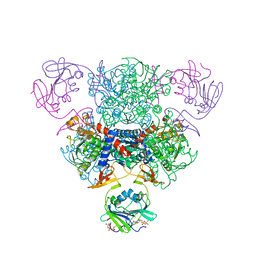 | | The Structure of Wild-Type E. Coli Aspartate Transcarbamoylase in Complex with Novel T State Inhibitors at 2.25 Resolution | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Heng, S, Stieglitz, K.A, Eldo, J, Xia, J, Cardia, J.P, Kantrowitz, E.R. | | Deposit date: | 2006-02-09 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | T-state Inhibitors of E. coli Aspartate Transcarbamoylase that Prevent the Allosteric Transition.
Biochemistry, 45, 2006
|
|
2FZK
 
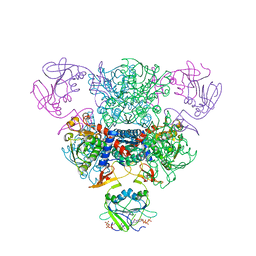 | | The Structure of Wild-Type E. Coli Aspartate Transcarbamoylase in Complex with Novel T State Inhibitors at 2.50 Resolution | | Descriptor: | 3,5-BIS[(PHOSPHONOACETYL)AMINO]BENZOIC ACID, Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, ... | | Authors: | Heng, S, Stieglitz, K.A, Eldo, J, Xia, J, Cardia, J.P, Kantrowitz, E.R. | | Deposit date: | 2006-02-09 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | T-state Inhibitors of E. coli Aspartate Transcarbamoylase that Prevent the Allosteric Transition.
Biochemistry, 45, 2006
|
|
2FZC
 
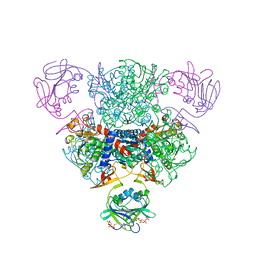 | | The Structure of Wild-Type E. Coli Aspartate Transcarbamoylase in Complex with Novel T State Inhibitors at 2.10 Resolution | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Heng, S, Stieglitz, K.A, Eldo, J, Xia, J, Cardia, J.P, Kantrowitz, E.R. | | Deposit date: | 2006-02-09 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | T-state Inhibitors of E. coli Aspartate Transcarbamoylase that Prevent the Allosteric Transition.
Biochemistry, 45, 2006
|
|
6NRW
 
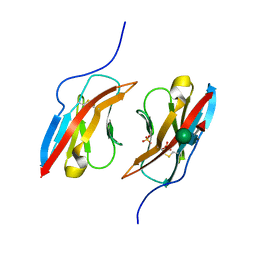 | | Crystal structure of Dpr1 IG1 bound to DIP-eta IG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cheng, S, Park, Y.J, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila.
Elife, 8, 2019
|
|
6NRX
 
 | | Crystal structure of DIP-eta IG1 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dpr-interacting protein eta, isoform B, ... | | Authors: | Cheng, S, Park, Y.J, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila.
Elife, 8, 2019
|
|
6NRQ
 
 | | Crystal structure of Dpr10 IG1 bound to DIP-alpha IG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Defective proboscis extension response 10, isoform A, ... | | Authors: | Cheng, S, Park, Y.J, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila.
Elife, 8, 2019
|
|
6NS1
 
 | | Crystal structure of DIP-gamma IG1+IG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Dpr-interacting protein gamma | | Authors: | Cheng, S, Park, Y.J, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila.
Elife, 8, 2019
|
|
8JKB
 
 | | Cryo-EM structure of KCTD5 in complex with Gbeta gamma subunits | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | | Authors: | Zheng, S, Jiang, W, Wang, W, Kong, Y. | | Deposit date: | 2023-06-01 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural basis for the ubiquitination of G protein beta gamma subunits by KCTD5/Cullin3 E3 ligase.
Sci Adv, 9, 2023
|
|
6PLL
 
 | | Crystal structure of the ZIG-8 IG1 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Zwei Ig domain protein zig-8 | | Authors: | Cheng, S, Ozkan, E. | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6ONB
 
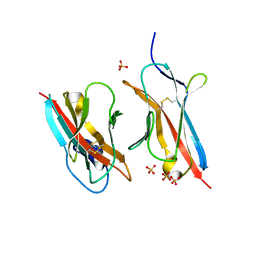 | | Crystal Structure of the ZIG-8-RIG-5 IG1-IG1 heterodimer, monoclinic form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NeuRonal IgCAM-5, ... | | Authors: | Cheng, S, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-04-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6ON9
 
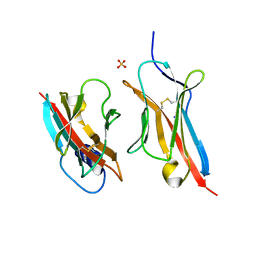 | | Crystal Structure of the ZIG-8-RIG-5 IG1-IG1 heterodimer, tetragonal form | | Descriptor: | NeuRonal IgCAM-5, SODIUM ION, SULFATE ION, ... | | Authors: | Cheng, S, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-04-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6ON6
 
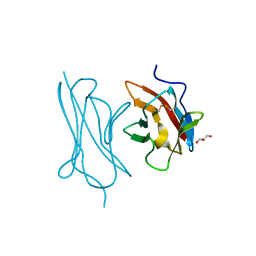 | | Crystal Structure of the RIG-5 IG1 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NeuRonal IgCAM-5 | | Authors: | Cheng, S, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-04-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5L2E
 
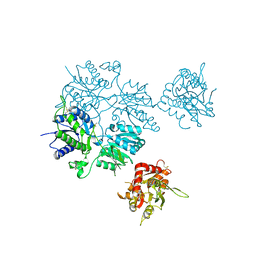 | |
4QMF
 
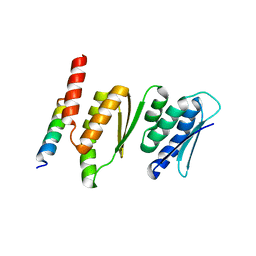 | | Structure of the Krr1 and Faf1 complex from Saccharomyces cerevisiae | | Descriptor: | KRR1 small subunit processome component, Protein FAF1 | | Authors: | Zheng, S, Ye, K. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Interaction between ribosome assembly factors Krr1 and Faf1 is essential for formation of small ribosomal subunit in yeast
J.Biol.Chem., 289, 2014
|
|
5KWR
 
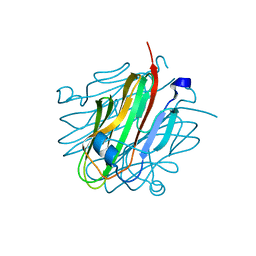 | | Crystal structure of rat Cerebellin-1 | | Descriptor: | Cerebellin-1, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Cheng, S, Ozkan, E. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Conformational Plasticity in the Transsynaptic Neurexin-Cerebellin-Glutamate Receptor Adhesion Complex.
Structure, 24, 2016
|
|
4RKH
 
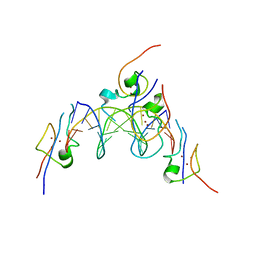 | | Structure of the MSL2 CXC domain bound with a specific MRE sequence | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*AP*TP*CP*TP*CP*GP*CP*TP*CP*AP*T)-3'), DNA (5'-D(*AP*TP*GP*AP*GP*CP*GP*AP*GP*AP*TP*GP*GP*AP*T)-3'), E3 ubiquitin-protein ligase msl-2, ... | | Authors: | Zheng, S, Ye, K. | | Deposit date: | 2014-10-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of X chromosome DNA recognition by the MSL2 CXC domain during Drosophila dosage compensation.
Genes Dev., 28, 2014
|
|
8CY8
 
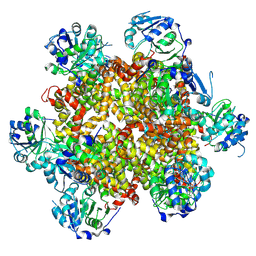 | |
1XNL
 
 | | ASLV fusion peptide | | Descriptor: | membrane protein gp37 | | Authors: | Cheng, S.F, Wu, C.W, Kantchev, E.A, Chang, D.K. | | Deposit date: | 2004-10-05 | | Release date: | 2005-01-11 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure and membrane interaction of the internal fusion peptide of avian sarcoma leukosis virus
Eur.J.Biochem., 271, 2004
|
|
5Z1G
 
 | | Structure of the Brx1 and Ebp2 complex | | Descriptor: | Ribosome biogenesis protein BRX1, SULFATE ION, rRNA-processing protein EBP2 | | Authors: | Zheng, S, Ye, K. | | Deposit date: | 2017-12-26 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate
Protein Cell, 10, 2019
|
|
4RKG
 
 | | Structure of the MSL2 CXC domain bound with a non-specific (GC)6 DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*C)-3'), E3 ubiquitin-protein ligase msl-2, ZINC ION | | Authors: | Zheng, S, Ye, K. | | Deposit date: | 2014-10-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of X chromosome DNA recognition by the MSL2 CXC domain during Drosophila dosage compensation.
Genes Dev., 28, 2014
|
|
5Z8O
 
 | |
2KGP
 
 | | Structural basis for stabilization of the tau pre-mRNA splicing regulatory element by Novantrone (Mitoxantrone) | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, RNA (25-MER) | | Authors: | Zheng, S, Chen, Y, Donahue, C.P, Wolfe, M.S, Varani, G. | | Deposit date: | 2009-03-13 | | Release date: | 2009-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for stabilization of the tau pre-mRNA splicing regulatory element by novantrone (mitoxantrone).
Chem.Biol., 16, 2009
|
|
6M8S
 
 | | Crystal structure of the KCTD12 H1 domain in complex with Gbeta1gamma2 subunits | | Descriptor: | BTB/POZ domain-containing protein KCTD12, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | | Authors: | Zheng, S, Kruse, A.C. | | Deposit date: | 2018-08-22 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Structural basis for KCTD-mediated rapid desensitization of GABABsignalling.
Nature, 567, 2019
|
|
2KE0
 
 | | Solution structure of peptidyl-prolyl cis-trans isomerase from Burkholderia pseudomallei | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Zheng, S, Leeper, T, Napuli, A, Nakazawa, S.H, Varani, G, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structure of a Burkholderia pseudomallei immunophilin-inhibitor complex reveals new approaches to antimicrobial development.
Biochem.J., 437, 2011
|
|
6CC4
 
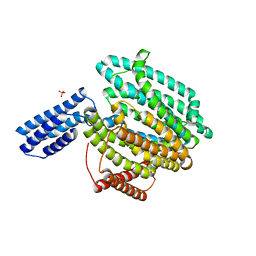 | | Structure of MurJ from Escherichia coli | | Descriptor: | PHOSPHATE ION, soluble cytochrome b562, lipid II flippase MurJ chimera | | Authors: | Zheng, S, Kruse, A.C. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and mutagenic analysis of the lipid II flippase MurJ fromEscherichia coli.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
