4REG
 
 | |
4L36
 
 | | Crystal structure of the cytochrome P450 enzyme TxtE | | Descriptor: | HEME B/C, IMIDAZOLE, Putative P450-like protein | | Authors: | Yu, F, Li, M.J, Xu, C.Y, Wang, Z.J, Zhou, H, Yang, M, Chen, Y.X, Tang, L, He, J.H. | | Deposit date: | 2013-06-05 | | Release date: | 2013-12-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural Insights into the Mechanism for Recognizing Substrate of the Cytochrome P450 Enzyme TxtE.
Plos One, 8, 2013
|
|
4PXG
 
 | |
4R6M
 
 | | Crystal Structure of DNA Duplex Containing Two Consecutive Mercury-mediated Base Pairs | | Descriptor: | DNA (5'-D(P*CP*GP*CP*GP*TP*TP*GP*TP*CP*C)-3'), DNA (5'-D(P*GP*GP*AP*CP*TP*TP*CP*GP*CP*G)-3'), MERCURY (II) ION | | Authors: | Xu, C.Y, Yu, F, Wang, L.H, Zhao, Y, Lin, T, Fan, C.H, He, J.H. | | Deposit date: | 2014-08-26 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.357 Å) | | Cite: | Crystal Structure of DNA Duplex Containing Two Consecutive Mercury-mediated Base Pairs
To be Published
|
|
4E5Y
 
 | |
4HY6
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ1 | | Descriptor: | 6,6-dimethyl-3-(trifluoromethyl)-1,5,6,7-tetrahydro-4H-indazol-4-one, Heat shock protein HSP 90-alpha | | Authors: | Yang, M, Li, J, He, J.H. | | Deposit date: | 2012-11-13 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ1
to be published
|
|
5XQD
 
 | | Crystal structure of Human Hsp90 with FS2 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]propanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Human Hsp90 with FS2
To Be Published
|
|
5XR5
 
 | | Crystal structure of Human Hsp90 with FS4 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]-2,2-dimethyl-propanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Human Hsp90 with FS4
To Be Published
|
|
5XRB
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-08 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5
To Be Published
|
|
5XQE
 
 | | Crystal structure of Human Hsp90 with FS3 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]-2-methyl-propanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of Human Hsp90 with FS3
To Be Published
|
|
5XRD
 
 | |
5XRE
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor JX1 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[4-(4-methoxyphenyl)-3-[5-(8-methylquinolin-5-yl)-2,4-bis(oxidanyl)phenyl]-1,2-oxazol-5-yl]ethanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-08 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor JX1
To Be Published
|
|
5XR9
 
 | | Crystal structure of Human Hsp90 with FS6 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-bromanyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]ethanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of Human Hsp90 with FS6
To Be Published
|
|
6A9W
 
 | | Structure of the bifunctional DNA primase-polymerase from phage NrS-1 | | Descriptor: | Primase | | Authors: | Guo, H.J, Li, M.J, Wang, T.L, Wu, H, Zhou, H, Xu, C.Y, Liu, X.P, Yu, F, He, J.H. | | Deposit date: | 2018-07-16 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and biochemical studies of the bifunctional DNA primase-polymerase from phage NrS-1.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
5X4J
 
 | | The crystal structure of Pyrococcus furiosus RecJ (Zn-soaking) | | Descriptor: | CHLORIDE ION, Uncharacterized protein, ZINC ION | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5X4K
 
 | | The complex crystal structure of Pyrococcus furiosus RecJ and CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Uncharacterized protein, ZINC ION | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5X4H
 
 | | The crystal structure of Pyrococcus furiosus RecJ (wild-type) | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5X4I
 
 | | Pyrococcus furiosus RecJ (D83A, Mn-soaking) | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Uncharacterized protein | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5GM1
 
 | | Crystal structure of methyltransferase TleD complexed with SAH | | Descriptor: | O-methylransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yu, F, Li, M.J, Xu, C.Y, Zhou, H, Sun, B, Wang, Z.J, Xu, Q, Xie, M.Y, Zuo, G, Huang, P, Wang, Q.S, He, J.H. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal structure and enantioselectivity of terpene cyclization in SAM-dependent methyltransferase TleD
Biochem.J., 473, 2016
|
|
5GM2
 
 | | Crystal structure of methyltransferase TleD complexed with SAH and teleocidin A1 | | Descriptor: | (2S,5S)-9-[(3R)-3,7-dimethylocta-1,6-dien-3-yl]-5-(hydroxymethyl)-1-methyl-2-(propan-2-yl)-1,2,4,5,6,8-hexahydro-3H-[1,4]diazonino[7,6,5-cd]indol-3-one, O-methylransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yu, F, Li, M.J, Xu, C.Y, Zhou, H, Sun, B, Wang, Z.J, Xu, Q, Xie, M.Y, Zuo, G, Huang, P, Wang, Q.S, He, J.H. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and enantioselectivity of terpene cyclization in SAM-dependent methyltransferase TleD
Biochem.J., 473, 2016
|
|
5CF0
 
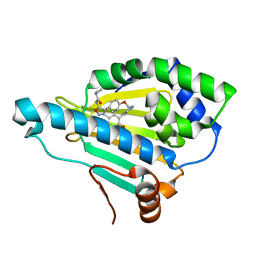 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ6 | | Descriptor: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(isoquinolin-4-yl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl}cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2015-07-08 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | FS23 binds to the N-terminal domain of human Hsp90: A novel small inhibitor for Hsp90
Nucl.Sci.Tech., 26, 2015
|
|
6JOQ
 
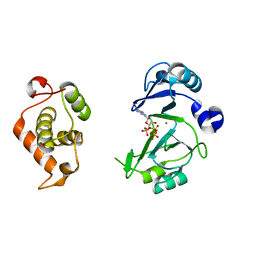 | | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+ complex provide molecular mechanisms for substrate specificity | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Primase | | Authors: | Guo, H.J, Li, M.J, Wu, H, Yu, F, He, J.H. | | Deposit date: | 2019-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+complex provide molecular mechanisms for substrate specificity.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
6IDE
 
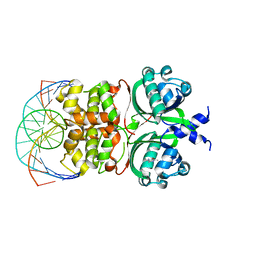 | | Crystal structure of the Vibrio cholera VqmA-Ligand-DNA complex provides molecular mechanisms for drug design | | Descriptor: | 3,5-dimethylpyrazin-2-ol, DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*AP*AP*TP*CP*CP*CP*CP*CP*CP*T)-3'), DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*TP*TP*TP*CP*CP*CP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Li, B, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2018-09-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of theVibrio choleraeVqmA-ligand-DNA complex provides insight into ligand-binding mechanisms relevant for drug design.
J. Biol. Chem., 294, 2019
|
|
6KKL
 
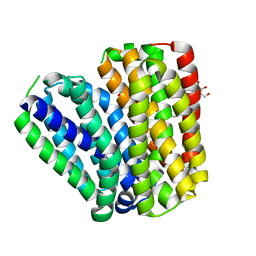 | | Crystal structure of Drug:Proton Antiporter-1 (DHA1) Family SotB, in the inward conformation (H115N mutant) | | Descriptor: | Sugar efflux transporter, nonyl beta-D-glucopyranoside | | Authors: | Xiao, Q.J, Sun, B, Zuo, Y.X, Guo, L, He, J.H, Deng, D. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Visualizing the nonlinear changes of a drug-proton antiporter from inward-open to occluded state.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
6KJU
 
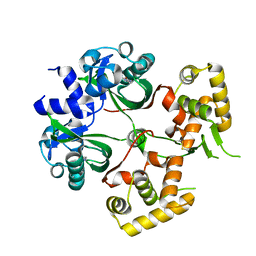 | | Huge conformation shift of Vibrio cholerae VqmA dimer in the absence of target DNA provides insight into DNA-binding mechanisms of LuxR-type receptors | | Descriptor: | 3,5-dimethylpyrazin-2-ol, Helix-turn-helix transcriptional regulator | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Wang, W.W, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Large conformation shifts of Vibrio cholerae VqmA dimer in the absence of target DNA provide insight into DNA-binding mechanisms of LuxR-type receptors.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
