3B1T
 
 | | Crystal structure of human peptidylarginine deiminase 4 in complex with o-Cl-amidine | | Descriptor: | 2-{[(2S)-1-amino-5-{[(1Z)-2-chloroethanimidoyl]amino}-1-oxopentan-2-yl]carbamoyl}benzoic acid, CALCIUM ION, Protein-arginine deiminase type-4, ... | | Authors: | Causey, C.P, Jones, J.E, Slack, J.L, Kamei, D, Jones Jr, L.E, Subramanian, V, Knuckley, B, Ebrahimi, P, Chumanevich, A.A, Luo, Y, Hashimoto, H, Shimizu, T, Sato, M, Hofseth, L.J, Thompson, P.R. | | Deposit date: | 2011-07-13 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Development of N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-fluoro-1-iminoethyl)-l-ornithine Amide (o-F-amidine) and N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-chloro-1-iminoethyl)-l-ornithine Amide (o-Cl-amidine) As Second Generation Protein Arginine Deiminase (PAD) Inhibitors
J.Med.Chem., 54, 2011
|
|
5H6K
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | Descriptor: | 1,2-ETHANEDIOL, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6N
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1, autoinhibitory form | | Descriptor: | Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6L
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 in complex with beta-NAD+ | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6M
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | Descriptor: | 1,2-ETHANEDIOL, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
7FI3
 
 | | Archaeal oligopeptide permease A (OppA) from Thermococcus kodakaraensis in complex with an endogenous pentapeptide | | Descriptor: | ABC-type dipeptide/oligopeptide transport system, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Yokoyama, H, Kamei, N, Konishi, K, Hara, K, Hashimoto, H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for peptide recognition by archaeal oligopeptide permease A.
Proteins, 90, 2022
|
|
5H6J
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 in complex with beta-NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
8FJM
 
 | |
8GNN
 
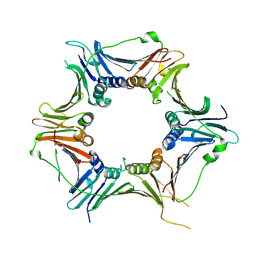 | | Crystal structure of the human RAD9-RAD1-HUS1-RAD17 complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Cell cycle checkpoint protein RAD17, ... | | Authors: | Hara, K, Nagata, K, Iida, N, Hashimoto, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | The 9-1-1 DNA clamp subunit RAD1 forms specific interactions with clamp loader RAD17, revealing functional implications for binding-protein RHINO.
J.Biol.Chem., 299, 2023
|
|
5FHI
 
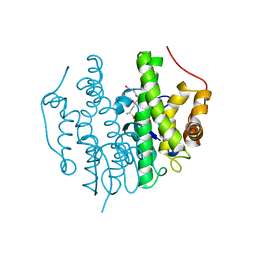 | | Crystallographic structure of PsoE without Co | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, putative | | Authors: | Hara, K, Hashimoto, H, Yamamoto, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2015-12-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Oxidative trans to cis Isomerization of Olefins in Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
5F8B
 
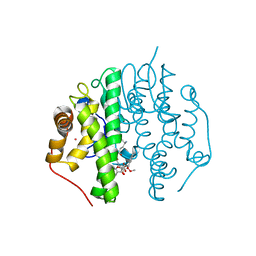 | | Crystallographic Structure of PsoE with Co | | Descriptor: | (5~{S},8~{S},9~{R})-2-[(~{E})-hex-1-enyl]-8-methoxy-3-methyl-9-oxidanyl-8-(phenylcarbonyl)-1-oxa-7-azaspiro[4.4]non-2-ene-4,6-dione, COBALT (II) ION, GLUTATHIONE, ... | | Authors: | Hara, K, Hashimoto, H, Yamamoto, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2015-12-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Oxidative trans to cis Isomerization of Olefins in Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
4XZF
 
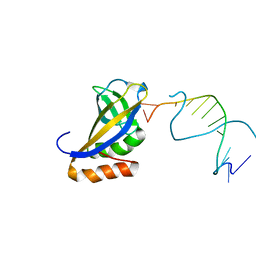 | | Crystal structure of HIRAN domain of human HLTF in complex with DNA | | Descriptor: | (DA)(DC)(DC)(DG)(DC)(DC)(DG)(DG)(DG)(DT)(DG)(DC)(DC), Helicase-like transcription factor | | Authors: | Hishiki, A, Hashimoto, H. | | Deposit date: | 2015-02-04 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure of a Novel DNA-binding Domain of Helicase-like Transcription Factor (HLTF) and Its Functional Implication in DNA Damage Tolerance
J.Biol.Chem., 290, 2015
|
|
5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | Authors: | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2015-11-25 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
1WN7
 
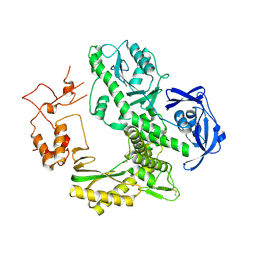 | | Crystal structure of archaeal family B DNA polymerase mutant | | Descriptor: | Family B DNA Polymerase, GLYCEROL, NICKEL (II) ION | | Authors: | Kuroita, T, Matsumura, H, Yokota, N, Hashimoto, H, Imanaka, T, Inoue, T, Kai, Y. | | Deposit date: | 2004-07-28 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Mechanism for Coordination of Proofreading and Polymerase Activities in Archaeal DNA Polymerases
J.Mol.Biol., 351, 2005
|
|
5XPT
 
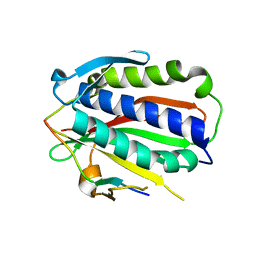 | |
5XPU
 
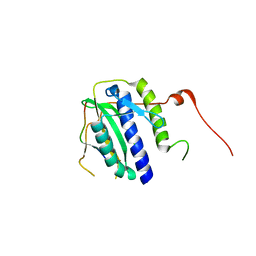 | |
7CP7
 
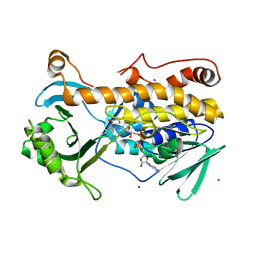 | | Crystal structure of FqzB, native proteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2020-08-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
8FJN
 
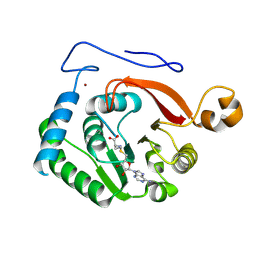 | |
7CP6
 
 | | Crystal structure of FqzB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase, ... | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2020-08-06 | | Release date: | 2020-12-30 | | Last modified: | 2021-01-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
6J8Y
 
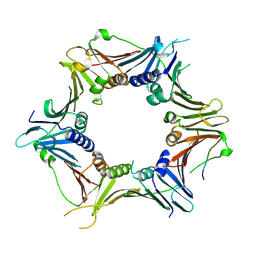 | | Crystal structure of the human RAD9-HUS1-RAD1-RHINO complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Hara, K, Iida, N, Sakurai, H, Hashimoto, H. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RAD9-RAD1-HUS1 checkpoint clamp bound to RHINO sheds light on the other side of the DNA clamp.
J.Biol.Chem., 295, 2020
|
|
6MR4
 
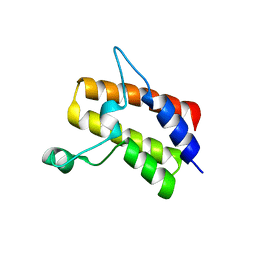 | | Crystal structure of the Sth1 bromodomain from S.cerevisiae | | Descriptor: | Nuclear protein STH1/NPS1 | | Authors: | Seo, H.S, Hashimoto, H, Krolak, A, Debler, E.W, Blus, B.J. | | Deposit date: | 2018-10-11 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Substrate Affinity and Specificity of the ScSth1p Bromodomain Are Fine-Tuned for Versatile Histone Recognition.
Structure, 27, 2019
|
|
1UHN
 
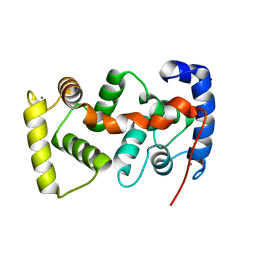 | | The crystal structure of the calcium binding protein AtCBL2 from Arabidopsis thaliana | | Descriptor: | CALCIUM ION, calcineurin B-like protein 2 | | Authors: | Nagae, M, Nozawa, A, Koizumi, N, Sano, H, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2003-07-07 | | Release date: | 2003-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Novel Calcium-binding Protein AtCBL2 from Arabidopsis thaliana
J.Biol.Chem., 278, 2003
|
|
1WDA
 
 | | Crystal structure of human peptidylarginine deiminase type4 (PAD4) in complex with benzoyl-L-arginine amide | | Descriptor: | CALCIUM ION, N-[(E)-2-AMINO-1-(3-{[AMINO(IMINO)METHYL]AMINO}PROPYL)-2-HYDROXYVINYL]BENZAMIDE, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Hashimoto, H, Shimizu, T, Nakashima, K, Yamada, M, Sato, M. | | Deposit date: | 2004-05-12 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Ca(2+)-induced activation of human PAD4
Nat.Struct.Mol.Biol., 11, 2004
|
|
1WD9
 
 | | Calcium bound form of human peptidylarginine deiminase type4 (PAD4) | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type IV, SULFATE ION | | Authors: | Arita, K, Hashimoto, H, Shimizu, T, Nakashima, K, Yamada, M, Sato, M. | | Deposit date: | 2004-05-12 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Ca(2+)-induced activation of human PAD4
Nat.Struct.Mol.Biol., 11, 2004
|
|
7YBD
 
 | | Crystal structure of sliding DNA clamp of Clostridioides difficile | | Descriptor: | Beta sliding clamp, TRIETHYLENE GLYCOL | | Authors: | Hishiki, A, Okazaki, S, Hara, K, Hashimoto, H. | | Deposit date: | 2022-06-29 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of the sliding DNA clamp from the Gram-positive anaerobic bacterium Clostridioides difficile.
J.Biochem., 173, 2022
|
|
