6TDH
 
 | | Crystal structure of Aspergillus fumigatus Glucosamine-6-phosphate N-acetyltransferase 1 in complex with compound 1 | | Descriptor: | 3-(2-chlorophenyl)-4~{H}-1,2,4-triazole, ACETYL COENZYME *A, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Raimi, O.G, Stanley, M, Lockhart, D. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Targeting a critical step in fungal hexosamine biosynthesis.
J.Biol.Chem., 295, 2020
|
|
6TDG
 
 | | Crystal structure of Aspergillus fumigatus Glucosamine-6-phosphate N-acetyltransferase 1 in complex with compound 2 | | Descriptor: | 2-chloranyl-3-(4~{H}-1,2,4-triazol-3-yl)aniline, ACETYL COENZYME *A, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Raimi, O.G, Stanley, M, Lockhart, D. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Targeting a critical step in fungal hexosamine biosynthesis.
J.Biol.Chem., 295, 2020
|
|
6SPT
 
 | | High resolution crystal structure of N-terminal domain of PEX14 from Trypanosoma brucei in complex with the fist compound with sub-micromolar trypanocidal activity | | Descriptor: | 5-[(4-methoxynaphthalen-1-yl)methyl]-1-[2-[(2-methyl-1-oxidanyl-propan-2-yl)amino]ethyl]-~{N}-(naphthalen-1-ylmethyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridine-3-carboxamide, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Napolitano, V, Dawidowski, M, Kalel, V.C, Fino, R, Emmanouilidis, L, Lenhart, D, Ostertag, M, Kaiser, M, Kolonko, M, Schilebs, W, Maser, P, Tetko, I, Hadian, K, Plettenburg, O, Erdmann, R, Sattler, M, Popowicz, G.M, Dubin, G. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Activity Relationship in Pyrazolo[4,3-c]pyridines, First Inhibitors of PEX14-PEX5 Protein-Protein Interaction with Trypanocidal Activity.
J.Med.Chem., 63, 2020
|
|
1MDR
 
 | | THE ROLE OF LYSINE 166 IN THE MECHANISM OF MANDELATE RACEMASE FROM PSEUDOMONAS PUTIDA: MECHANISTIC AND CRYSTALLOGRAPHIC EVIDENCE FOR STEREOSPECIFIC ALKYLATION BY (R)-ALPHA-PHENYLGLYCIDATE | | Descriptor: | ATROLACTIC ACID (2-PHENYL-LACTIC ACID), MAGNESIUM ION, MANDELATE RACEMASE | | Authors: | Landro, J.A, Gerlt, J.A, Kozarich, J.W, Koo, C.W, Shah, V.J, Kenyon, G.L, Neidhart, D.J, Fujita, S, Petsko, G.A. | | Deposit date: | 1993-11-19 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of lysine 166 in the mechanism of mandelate racemase from Pseudomonas putida: mechanistic and crystallographic evidence for stereospecific alkylation by (R)-alpha-phenylglycidate.
Biochemistry, 33, 1994
|
|
2EPL
 
 | | N-acetyl-B-D-glucosaminidase (GCNA) from Streptococcus gordonii | | Descriptor: | GLYCEROL, N-acetyl-beta-D-glucosaminidase, SULFATE ION | | Authors: | Langley, D.B, Harty, D.W.S, Guss, J.M, Collyer, C.A. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
2EPM
 
 | | N-acetyl-B-D-glucoasminidase (GCNA) from Stretococcus gordonii | | Descriptor: | GLYCEROL, MERCURY (II) ION, N-acetyl-beta-D-glucosaminidase, ... | | Authors: | Langley, D.B, Harty, D.W.S, Guss, J.M, Collyer, C.A. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
4CAV
 
 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a benzofuran ligand R0-09-4879 | | Descriptor: | 3-[[3-methyl-2-[[2,3,4-tris(fluoranyl)phenoxy]methyl]-1-benzofuran-4-yl]oxy]-N-(pyridin-3-ylmethyl)propan-1-amine, CHLORIDE ION, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Robinson, D.A, Fang, W, Raimi, O.G, Blair, D.E, Harrison, J, Ruda, G.F, Lockhart, D.E.A, Torrie, L.S, Wyatt, P.G, Gilbert, I.H, Van Aalten, D.M.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | N-Myristoyltransferase is a Cell Wall Target in Aspergillus Fumigatus.
Acs Chem.Biol., 10, 2015
|
|
4CAW
 
 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a pyrazole sulphonamide ligand | | Descriptor: | 2,6-DICHLORO-4-(2-PIPERAZIN-1-YLPYRIDIN-4-YL)-N-(1,5-DIMETHYL,3-ISOBUTYL-1H-PYRAZOL-4-YL)BENZENESULFONAMIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Fang, W, Raimi, O.G, Blair, D.E, Harrison, J, Ruda, G.F, Lockhart, D.E.A, Torrie, L.S, Wyatt, P.G, Gilbert, I.H, Van Aalten, D.M.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | N-Myristoyltransferase is a Cell Wall Target in Aspergillus Fumigatus.
Acs Chem.Biol., 10, 2015
|
|
4CAX
 
 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a pyrazole sulphonamide ligand (DDD85646) | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Raimi, O.G, Robinson, D.A, Fang, W, Blair, D.E, Harrison, J, Ruda, G.F, Lockhart, D.E.A, Torrie, L.S, Wyatt, P.G, Gilbert, I.H, Van Aalten, D.M.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N-Myristoyltransferase is a Cell Wall Target in Aspergillus Fumigatus.
Acs Chem.Biol., 10, 2015
|
|
4BBA
 
 | | Crystal structure of glucokinase regulatory protein complexed to phosphate | | Descriptor: | GLUCOKINASE REGULATORY PROTEIN, PHOSPHATE ION | | Authors: | Pautsch, A, Stadler, N, Loehle, A, Lenter, M, Rist, W, Berg, A, Glocker, L, Nar, H, Reinhart, D, Heckel, A, Schnapp, G, Kauschke, S.G. | | Deposit date: | 2012-09-21 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Glucokinase Regulatory Protein.
Biochemistry, 52, 2013
|
|
1ILO
 
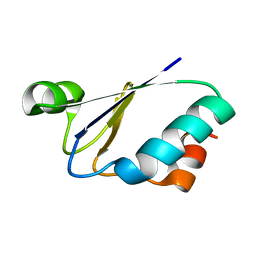 | | NMR structure of a thioredoxin, MtH895, from the archeon Methanobacterium thermoautotrophicum strain delta H. | | Descriptor: | conserved hypothetical protein MtH895 | | Authors: | Bhattacharyya, S, Habibi-Nazhad, B, Slupsky, C.M, Sykes, B.D, Wishart, D.S, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-05-08 | | Release date: | 2001-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Identification of a novel archaebacterial thioredoxin: determination of function through structure.
Biochemistry, 41, 2002
|
|
1U9Q
 
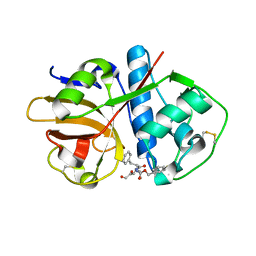 | | Crystal structure of cruzain bound to an alpha-ketoester | | Descriptor: | [1-(1-METHYL-4,5-DIOXO-PENT-2-ENYLCARBAMOYL)-2-PHENYL-ETHYL]-CARBAMIC ACID BENZYL ESTER, cruzipain | | Authors: | Lange, M, Weston, S.G, Cheng, H, Culliane, M, Fiorey, M.M, Grisostomi, C, Hardy, L.W, Hartstough, D.S, Pallai, P.V, Tilton, R.F, Baldino, C.M, Brinen, L.S, Engel, J.C, Choe, Y, Price, M.S, Craik, C.S. | | Deposit date: | 2004-08-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of alpha-keto-based inhibitors of cruzain, a cysteine protease implicated in Chagas disease
Bioorg.Med.Chem., 13, 2005
|
|
1NHO
 
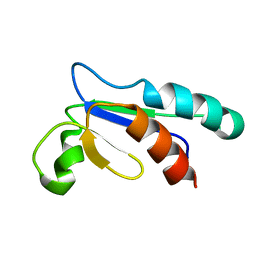 | | Structural and Functional characterization of a Thioredoxin-like Protein from Methanobacterium thermoautotrophicum | | Descriptor: | Probable Thioredoxin | | Authors: | Amegbey, G.Y, Monzavi, H, Habibi-Nazhad, B, Bhattacharyya, S, Wishart, D.S. | | Deposit date: | 2002-12-19 | | Release date: | 2003-08-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of a Thioredoxin-like Protein (Mt0807) from Methanobacterium thermoautotrophicum
Biochemistry, 42, 2003
|
|
2LH8
 
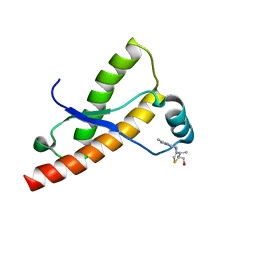 | | Syrian hamster prion protein with thiamine | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Major prion protein | | Authors: | Perez-Pineiro, R, Bjorndahl, T.C, Berjanskii, M, Hau, D, Li, L, Huang, A, Lee, R, Gibbs, E, Ladner, C, Wei Dong, Y, Abera, A, Cashman, N.R, Wishart, D. | | Deposit date: | 2011-08-05 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The prion protein binds thiamine.
Febs J., 278, 2011
|
|
1Z9V
 
 | | Solution Structure of MTH0776 from Methanobacterium thermoautotrophicum (strain H) | | Descriptor: | conserved hypothetical protein MTH0776 | | Authors: | Amegbey, G, Stothard, P, Iakounine, A, Yee, A, Arrowsmith, C.H, Wishart, D.S. | | Deposit date: | 2005-04-04 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of MTH0776 from Methanobacterium Thermoautotrophicum.
J.Biomol.Nmr, 33, 2005
|
|
1VJY
 
 | | Crystal Structure of a Naphthyridine Inhibitor of Human TGF-beta Type I Receptor | | Descriptor: | 2-[5-(6-METHYLPYRIDIN-2-YL)-2,3-DIHYDRO-1H-PYRAZOL-4-YL]-1,5-NAPHTHYRIDINE, TGF-beta receptor type I | | Authors: | Gellibert, F, Woolven, J, Fouchet, M.-H, Mathews, N, Goodland, H, Lovegrove, V, Laroze, A, Nguyen, V.-L, Sautet, S, Wang, R, Janson, C, Smith, W, Krysa, G, Boullay, V, de Gouville, A.-C, Huet, S, Hartley, D. | | Deposit date: | 2004-04-07 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of 1,5-Naphthyridine Derivatives as a Novel Series of Potent and Selective TGF-beta Type I Receptor Inhibitors.
J.Med.Chem., 47, 2004
|
|
1RY3
 
 | | NMR Solution Structure of the Precursor for Carnobacteriocin B2, an Antimicrobial Peptide from Carnobacterium piscicola | | Descriptor: | Bacteriocin carnobacteriocin B2 | | Authors: | Sprules, T, Kawulka, K.E, Gibbs, A.C, Wishart, D.S, Vederas, J.C. | | Deposit date: | 2003-12-19 | | Release date: | 2004-05-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the precursor for carnobacteriocin B2, an antimicrobial peptide from Carnobacterium piscicola.
Eur.J.Biochem., 271, 2004
|
|
1RYJ
 
 | | Solution NMR Structure of Protein Mth1743 from Methanobacterium thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH1743_1_70; Northeast Structural Genomics Consortium Target TT526. | | Descriptor: | unknown | | Authors: | Yee, A, Chang, X, Pineda-Lucena, A, Wu, B, Semesi, A, Le, B, Ramelot, T, Lee, G.M, Bhattacharyya, S, Gutierrez, P, Denisov, A, Lee, C.H, Cort, J.R, Kozlov, G, Liao, J, Finak, G, Chen, L, Wishart, D, Lee, W, McIntosh, L.P, Gehring, K, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | AN NMR APPROACH TO STRUCTURAL PROTEOMICS
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1CW5
 
 | | SOLUTION STRUCTURE OF CARNOBACTERIOCIN B2 | | Descriptor: | TYPE IIA BACTERIOCIN CARNOBACTERIOCIN B2 | | Authors: | Wang, Y, Henz, M.E, Gallagher, N.L.F, Chai, S, Yan, L.Z, Gibbs, A.C, Stiles, M.E, Wishart, D.S, Vederas, J.C. | | Deposit date: | 1999-08-25 | | Release date: | 1999-09-07 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of carnobacteriocin B2 and implications for structure-activity relationships among type IIa bacteriocins from lactic acid bacteria.
Biochemistry, 38, 1999
|
|
2B0F
 
 | |
1CW6
 
 | | REFINED SOLUTION STRUCTURE OF LEUCOCIN A | | Descriptor: | TYPE IIA BACTERIOCIN LEUCOCIN A | | Authors: | Wang, Y, Henz, M.E, Gallagher, N.L.F, Chai, S, Yan, L.Z, Gibbs, A.C, Stiles, M.E, Wishart, D.S, Vederas, J.C. | | Deposit date: | 1999-08-25 | | Release date: | 1999-09-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of carnobacteriocin B2 and implications for structure-activity relationships among type IIa bacteriocins from lactic acid bacteria.
Biochemistry, 38, 1999
|
|
1DE1
 
 | |
1DE2
 
 | |
2IN2
 
 | |
