4MEM
 
 | | Crystal Structure of the rat USP11 DUSP-UBL domains | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 11 | | Authors: | Harper, S, Gratton, H.E, Cornaciu, I, Oberer, M, Scott, D.J, Emsley, J, Dreveny, I. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and Catalytic Regulatory Function of Ubiquitin Specific Protease 11 N-Terminal and Ubiquitin-like Domains.
Biochemistry, 53, 2014
|
|
4MEL
 
 | | Crystal Structure of the human USP11 DUSP-UBL domains | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 11 | | Authors: | Harper, S, Gratton, H.E, Cornaciu, I, Oberer, M, Scott, D.J, Emsley, J, Dreveny, I. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structure and Catalytic Regulatory Function of Ubiquitin Specific Protease 11 N-Terminal and Ubiquitin-like Domains.
Biochemistry, 53, 2014
|
|
3T9L
 
 | | Structure of N-terminal DUSP-UBL domains of human USP15 | | Descriptor: | SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Harper, S, Besong, T.M.D, Emsley, J, Scott, D.J, Dreveny, I. | | Deposit date: | 2011-08-03 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the USP15 N-Terminal Domains: A beta-Hairpin Mediates Close Association between the DUSP and UBL Domains
Biochemistry, 50, 2011
|
|
3LBX
 
 | | Crystal Structure of the Erythrocyte Spectrin Tetramerization Domain Complex | | Descriptor: | Spectrin alpha chain, erythrocyte, Spectrin beta chain | | Authors: | Ipsaro, J.J, Harper, S.L, Messick, T.E, Marmorstein, R, Mondragon, A, Speicher, D.W. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and functional interpretation of the erythrocyte spectrin tetramerization domain complex.
Blood, 115, 2010
|
|
1MJB
 
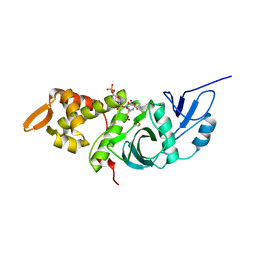 | | Crystal structure of yeast Esa1 histone acetyltransferase E338Q mutant complexed with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, Esa1 protein | | Authors: | Yan, Y, Harper, S, Speicher, D, Marmorstein, R. | | Deposit date: | 2002-08-27 | | Release date: | 2002-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.
Nat.Struct.Biol., 9, 2002
|
|
3COQ
 
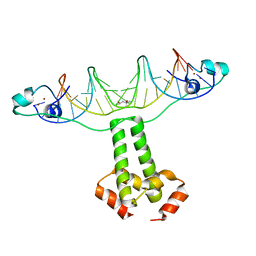 | | Structural Basis for Dimerization in DNA Recognition by Gal4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*DAP*DCP*DCP*DGP*DGP*DAP*DGP*DGP*DAP*DCP*DAP*DGP*DTP*DCP*DCP*DTP*DCP*DCP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DGP*DGP*DAP*DGP*DGP*DAP*DCP*DTP*DGP*DTP*DCP*DCP*DTP*DCP*DCP*DGP*DG)-3'), ... | | Authors: | Hong, M, Fitzgerald, M.X, Harper, S, Luo, C, Speicher, D.W. | | Deposit date: | 2008-03-29 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dimerization in DNA recognition by gal4.
Structure, 16, 2008
|
|
5JPZ
 
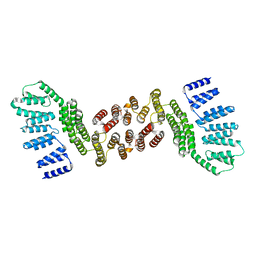 | | Crystal structure of HAT domain of human Squamous Cell Carcinoma Antigen Recognized By T Cells 3, SART3 (TIP110) | | Descriptor: | Squamous cell carcinoma antigen recognized by T-cells 3 | | Authors: | Grazette, A, Harper, S, Emsley, J, Layfield, R, Dreveny, I. | | Deposit date: | 2016-05-04 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.045 Å) | | Cite: | unpublished
To Be Published
|
|
3NWS
 
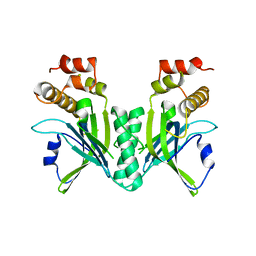 | | Crystal structure of the N-terminal domain of the yeast telomere-binding and telomerase regulatory protein Cdc13 | | Descriptor: | Cell division control protein 13 | | Authors: | Mitchell, M.T, Smith, J.S, Mason, M, Harper, S, Speicher, D.W, Johnson, F.B, Skordalakes, E. | | Deposit date: | 2010-07-10 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cdc13 N-terminal dimerization, DNA binding, and telomere length regulation.
Mol.Cell.Biol., 30, 2010
|
|
3NWT
 
 | | Crystal structure of the N-terminal domain of the yeast telomere-binding and telomerase regulatory protein Cdc13 | | Descriptor: | Cell division control protein 13 | | Authors: | Mitchell, M.T, Smith, J.S, Mason, M, Harper, S, Speicher, D.W, Johnson, F.B, Skordalakes, E. | | Deposit date: | 2010-07-10 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cdc13 N-terminal dimerization, DNA binding, and telomere length regulation.
Mol.Cell.Biol., 30, 2010
|
|
6RPA
 
 | | Crystal structure of the T-cell receptor NYE_S2 bound to HLA A2*01-SLLMWITQV | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Coles, C.H, Mulvaney, R, Malla, S, Lloyd, A, Smith, K, Chester, F, Knox, A, Stacey, A.R, Dukes, J, Baston, E, Griffin, S, Vuidepot, A, Jakobsen, B.K, Harper, S. | | Deposit date: | 2019-05-14 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | TCRs with Distinct Specificity Profiles Use Different Binding Modes to Engage an Identical Peptide-HLA Complex.
J Immunol., 204, 2020
|
|
6RP9
 
 | | Crystal structure of the T-cell receptor NYE_S3 bound to HLA A2*01-SLLMWITQV | | Descriptor: | Beta-2-microglobulin, Cancer/testis antigen 1, HLA class I histocompatibility antigen, ... | | Authors: | Coles, C.H, Mulvaney, R, Malla, S, Lloyd, A, Smith, K, Chester, F, Knox, A, Stacey, A.R, Dukes, J, Baston, E, Griffin, S, Vuidepot, A, Jakobsen, B.K, Harper, S. | | Deposit date: | 2019-05-14 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | TCRs with Distinct Specificity Profiles Use Different Binding Modes to Engage an Identical Peptide-HLA Complex.
J Immunol., 204, 2020
|
|
6RPB
 
 | | Crystal structure of the T-cell receptor NYE_S1 bound to HLA A2*01-SLLMWITQV | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Coles, C.H, Mulvaney, R, Malla, S, Lloyd, A, Smith, K, Chester, F, Knox, A, Stacey, A.R, Dukes, J, Baston, E, Griffin, S, Vuidepot, A, Jakobsen, B.K, Harper, S. | | Deposit date: | 2019-05-14 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | TCRs with Distinct Specificity Profiles Use Different Binding Modes to Engage an Identical Peptide-HLA Complex.
J Immunol., 204, 2020
|
|
4L4S
 
 | | Structural characterisation of the NADH binary complex of human lactate dehydrogenase M isozyme | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase A chain | | Authors: | Dempster, S, Harper, S, Moses, J.E, Dreveny, I. | | Deposit date: | 2013-06-09 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural characterization of the apo form and NADH binary complex of human lactate dehydrogenase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4L4R
 
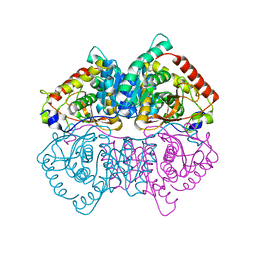 | | Structural Characterisation of the Apo-form of Human Lactate Dehydrogenase M Isozyme | | Descriptor: | L-lactate dehydrogenase A chain | | Authors: | Dempster, S, Harper, S, Moses, J.E, Dreveny, I. | | Deposit date: | 2013-06-09 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the apo form and NADH binary complex of human lactate dehydrogenase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6TRO
 
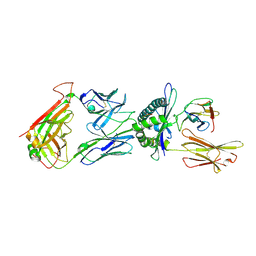 | | Crystal structure of the T-cell receptor GVY01 bound to HLA A2*01-GVYDGREHTV | | Descriptor: | Beta-2-microglobulin, MAGE-A4 peptide (amino acids 230-239), MHC class I antigen, ... | | Authors: | Coles, C.H, McMurran, C, Lloyd, A, Hibbert, L, Lupardus, P.J, Cole, D.K, Harper, S. | | Deposit date: | 2019-12-19 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | T cell receptor interactions with human leukocyte antigen govern indirect peptide selectivity for the cancer testis antigen MAGE-A4.
J.Biol.Chem., 295, 2020
|
|
6TRN
 
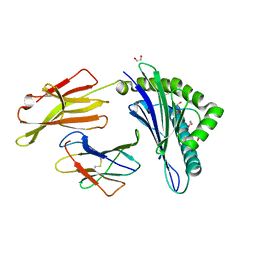 | | Crystal structure of HLA A2*01-AVYDGREHTV | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MAGE-A4 peptide (amino acids 230-239) variant, ... | | Authors: | Coles, C.H, McMurran, C, Lloyd, A, Hibbert, L, Lupardus, P.J, Cole, D.K, Harper, S. | | Deposit date: | 2019-12-19 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | T cell receptor interactions with human leukocyte antigen govern indirect peptide selectivity for the cancer testis antigen MAGE-A4.
J.Biol.Chem., 295, 2020
|
|
1LL8
 
 | |
4HCE
 
 | | Crystal structure of the telomeric Saccharomyces cerevisiae Cdc13 OB2 domain | | Descriptor: | Cell division control protein 13 | | Authors: | Mason, M, Wannat, J.J, Harper, S, Schultz, D.C, Speicher, D.W, Johnson, F.B, Skordalakes, E. | | Deposit date: | 2012-09-29 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cdc13 OB2 Dimerization Required for Productive Stn1 Binding and Efficient Telomere Maintenance.
Structure, 21, 2013
|
|
1MJ9
 
 | | Crystal structure of yeast Esa1(C304S) mutant complexed with Coenzyme A | | Descriptor: | COENZYME A, ESA1 PROTEIN, SODIUM ION | | Authors: | Yan, Y, Harper, S, Speicher, D, Marmorstein, R. | | Deposit date: | 2002-08-27 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.
Nat.Struct.Biol., 9, 2002
|
|
6SP1
 
 | | KEAP1 IN COMPLEX WITH COMPOUND 6 | | Descriptor: | (1~{S},2~{R})-2-[[(1~{S})-1-[[1,3-bis(oxidanylidene)isoindol-2-yl]methyl]-5-(2-hydroxyethyloxy)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]cyclohexane-1-carboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Ontoria, J.M, Biancofiore, I, Fezzardi, P, Torrente de Haro, E, Colarusso, S, Bianchi, E, Andreini, M, Patsilinakos, A, Summa, V, Pacifici, R, Munoz-Sanjuan, I, Park, L, Bresciani, A, Dominguez, C, Toledo-Sherman, L, Harper, S. | | Deposit date: | 2019-08-30 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Combined Peptide and Small-Molecule Approach toward Nonacidic THIQ Inhibitors of the KEAP1/NRF2 Interaction.
Acs Med.Chem.Lett., 11, 2020
|
|
6SP4
 
 | | KEAP1 IN COMPLEX WITH COMPOUND 23 | | Descriptor: | (1~{S},2~{R})-2-[[(1~{S})-1-[[1,3-bis(oxidanylidene)isoindol-2-yl]methyl]-5-(2-hydroxyethyloxy)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]cyclobutane-1-carboxamide, Kelch-like ECH-associated protein 1 | | Authors: | Ontoria, J.M, Biancofiore, I, Fezzardi, P, Torrente de Haro, E, Colarusso, S, Bianchi, E, Andreini, M, Patsilinakos, A, Summa, V, Pacifici, R, Munoz-Sanjuan, I, Park, L, Bresciani, A, Dominguez, C, Toledo-Sherman, L, Harper, S. | | Deposit date: | 2019-08-30 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Combined Peptide and Small-Molecule Approach toward Nonacidic THIQ Inhibitors of the KEAP1/NRF2 Interaction.
Acs Med.Chem.Lett., 11, 2020
|
|
1MJA
 
 | | Crystal structure of yeast Esa1 histone acetyltransferase domain complexed with acetyl coenzyme A | | Descriptor: | COENZYME A, Esa1 protein | | Authors: | Yan, Y, Harper, S, Speicher, D, Marmorstein, R. | | Deposit date: | 2002-08-27 | | Release date: | 2002-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.
Nat.Struct.Biol., 9, 2002
|
|
1HQ5
 
 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH S-(2-BORONOETHYL)-L-CYSTEINE, AN L-ARGININE ANALOGUE | | Descriptor: | ARGINASE 1, MANGANESE (II) ION, S-2-(BORONOETHYL)-L-CYSTEINE | | Authors: | Kim, N.N, Cox, J.D, Baggio, R.F, Emig, F.A, Mistry, S.K, Harper, S.L, Speicher, D.W, Morris Jr, S.M, Ash, D.E, Traish, A, Christianson, D.W. | | Deposit date: | 2000-12-14 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing erectile function: S-(2-boronoethyl)-L-cysteine binds to arginase as a transition state analogue and enhances smooth muscle relaxation in human penile corpus cavernosum.
Biochemistry, 40, 2001
|
|
