8YHR
 
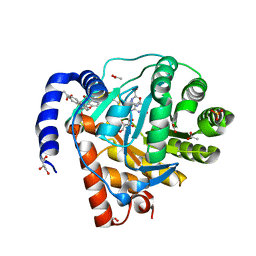 | | DHODH in complex with furocoumavirin | | Descriptor: | 4-methyl-8-[(S)-oxidanyl(phenyl)methyl]-9-phenyl-furo[2,3-h]chromen-2-one, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Hara, K, Okumura, H, Nakahara, M, Sato, M, Hashimoto, H, Osada, H, Watanabe, K. | | Deposit date: | 2024-02-28 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Analyses of Inhibition of Human Dihydroorotate Dehydrogenase by Antiviral Furocoumavirin.
Biochemistry, 63, 2024
|
|
8Y2S
 
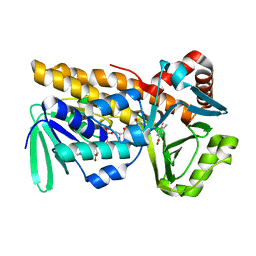 | | P-hydroxybenzoate hydroxylase complexed with 4-hydroxy-3-methylbenzoic acid | | Descriptor: | 3-methyl-4-oxidanyl-benzoic acid, 4-hydroxybenzoate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2024-01-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Enhancement of Flavin-Containing Monooxygenase through Machine Learning Methodology
Acs Catalysis, 14, 2024
|
|
8W7D
 
 | | Crystal structure of EcPPAT-FR901483 complex | | Descriptor: | Amidophosphoribosyltransferase, [(1S,3S,6S,7S,8R,9S)-6-[(4-methoxyphenyl)methyl]-3-(methylamino)-7-oxidanyl-5-azatricyclo[6.3.1.0^1,5]dodecan-9-yl] dihydrogen phosphate | | Authors: | Hara, K, Hashimoto, H, Nakahara, M, Sato, M, Watanabe, K. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Uncommon Arrangement of Self-resistance Allows Biosynthesis of de novo Purine Biosynthesis Inhibitor that Acts as an Immunosuppressor.
J.Am.Chem.Soc., 145, 2023
|
|
8GNN
 
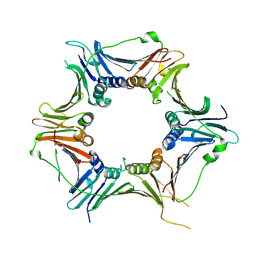 | | Crystal structure of the human RAD9-RAD1-HUS1-RAD17 complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Cell cycle checkpoint protein RAD17, ... | | Authors: | Hara, K, Nagata, K, Iida, N, Hashimoto, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | The 9-1-1 DNA clamp subunit RAD1 forms specific interactions with clamp loader RAD17, revealing functional implications for binding-protein RHINO.
J.Biol.Chem., 299, 2023
|
|
7BR0
 
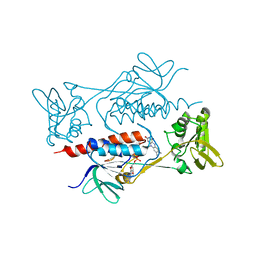 | | Crystal structure of AclR, a thioredoxin oxidoreductase fold protein carrying the CXXH catalytic motif | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyr_redox_2 domain-containing protein | | Authors: | Hara, K, Hashimoto, H, Maeda, N, Watanabe, K, Hertweck, C, Tsunematsu, Y. | | Deposit date: | 2020-03-26 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Specialized Flavoprotein Promotes Sulfur Migration and Spiroaminal Formation in Aspirochlorine Biosynthesis.
J.Am.Chem.Soc., 143, 2021
|
|
8WU8
 
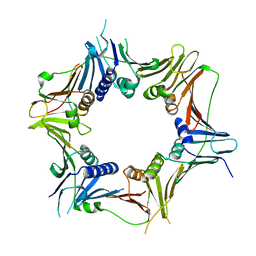 | | Crystal structure of the human RAD9-RAD1(F64A/M256A/F266A)-HUS1-RHINO(88-99) complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Hara, K, Nagata, K, Iida, N, Hashimoto, H. | | Deposit date: | 2023-10-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for intra- and intermolecular interactions on RAD9 subunit of 9-1-1 checkpoint clamp implies functional 9-1-1 regulation by RHINO.
J.Biol.Chem., 300, 2024
|
|
7CP6
 
 | | Crystal structure of FqzB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase, ... | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2020-08-06 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
7CP7
 
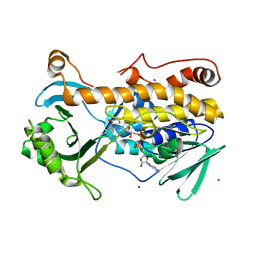 | | Crystal structure of FqzB, native proteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2020-08-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
5F8B
 
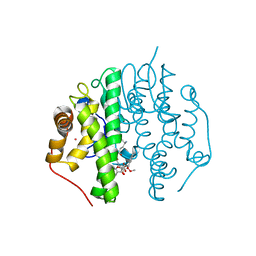 | | Crystallographic Structure of PsoE with Co | | Descriptor: | (5~{S},8~{S},9~{R})-2-[(~{E})-hex-1-enyl]-8-methoxy-3-methyl-9-oxidanyl-8-(phenylcarbonyl)-1-oxa-7-azaspiro[4.4]non-2-ene-4,6-dione, COBALT (II) ION, GLUTATHIONE, ... | | Authors: | Hara, K, Hashimoto, H, Yamamoto, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2015-12-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Oxidative trans to cis Isomerization of Olefins in Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
5XPT
 
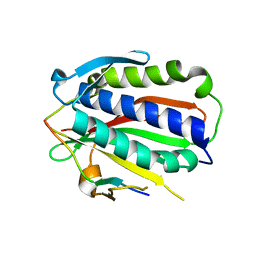 | |
5XPU
 
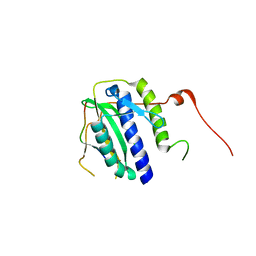 | |
5FHI
 
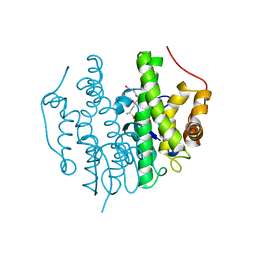 | | Crystallographic structure of PsoE without Co | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, putative | | Authors: | Hara, K, Hashimoto, H, Yamamoto, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2015-12-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Oxidative trans to cis Isomerization of Olefins in Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
6IGX
 
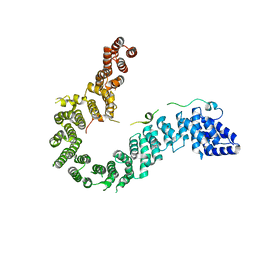 | | Crystal structure of human CAP-G in complex with CAP-H | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Condensin complex subunit 2, Condensin complex subunit 3 | | Authors: | Hara, K, Migita, T, Shimizu, K, Hashimoto, H. | | Deposit date: | 2018-09-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | Structural basis of HEAT-kleisin interactions in the human condensin I subcomplex.
Embo Rep., 20, 2019
|
|
5YY2
 
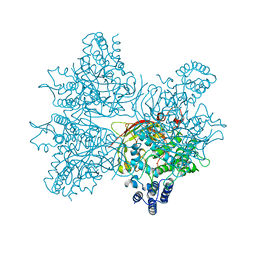 | | Crystal structure of AsqI with Zn | | Descriptor: | Uncharacterized protein AsqI, ZINC ION | | Authors: | Hara, K, Hashimoto, H, Kishimoto, S, Watanabe, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Enzymatic one-step ring contraction for quinolone biosynthesis.
Nat Commun, 9, 2018
|
|
5YY3
 
 | |
6KAW
 
 | | Crystal structure of CghA | | Descriptor: | CghA | | Authors: | Hara, K, Hashimoto, H, Yokoyama, M, Sato, M, Watanabe, K. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Catalytic mechanism and endo-to-exo selectivity reversion of an octalin-forming natural Diels-Alderase
Nat Catal, 2021
|
|
6KBC
 
 | | Crystal structure of CghA with Sch210972 | | Descriptor: | (2S)-3-[(2S,4E)-4-[[(1R,2S,4aR,6S,8R,8aS)-2-[(E)-but-2-en-2-yl]-6,8-dimethyl-1,2,4a,5,6,7,8,8a-octahydronaphthalen-1-yl]-oxidanyl-methylidene]-3,5-bis(oxidanylidene)pyrrolidin-2-yl]-2-methyl-2-oxidanyl-propanoic acid, CghA | | Authors: | Hara, K, Hashimoto, H, Maeda, N, Sato, M, Watanabe, K. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Catalytic mechanism and endo-to-exo selectivity reversion of an octalin-forming natural Diels-Alderase
Nat Catal, 2021
|
|
6KJI
 
 | | Crystal structure of PsoF with SAH | | Descriptor: | Dual-functional monooxygenase/methyltransferase psoF, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional and Structural Analyses oftrans C-Methyltransferase in Fungal Polyketide Biosynthesis.
Biochemistry, 58, 2019
|
|
6KJG
 
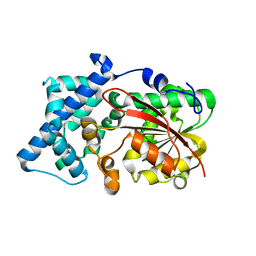 | | Crystal structure of PsoF | | Descriptor: | Dual-functional monooxygenase/methyltransferase psoF | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional and Structural Analyses oftrans C-Methyltransferase in Fungal Polyketide Biosynthesis.
Biochemistry, 58, 2019
|
|
6J8Y
 
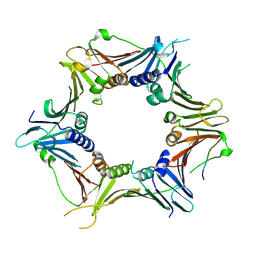 | | Crystal structure of the human RAD9-HUS1-RAD1-RHINO complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Hara, K, Iida, N, Sakurai, H, Hashimoto, H. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RAD9-RAD1-HUS1 checkpoint clamp bound to RHINO sheds light on the other side of the DNA clamp.
J.Biol.Chem., 295, 2020
|
|
4PJU
 
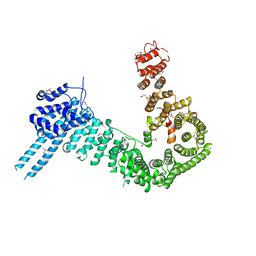 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1) | | Descriptor: | Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-12 | | Release date: | 2014-08-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4PJW
 
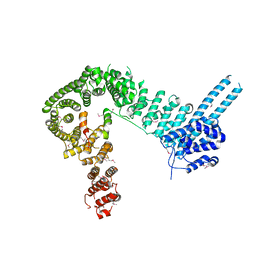 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1), with bound MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4PK7
 
 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1) with bound MES, native proteins | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-13 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6LPK
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 293 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
6LPJ
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 277 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
