8AEB
 
 | | SARS-CoV-2 Main Protease complexed with N-(pyridin-3-ylmethyl)thioformamide | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-(pyridin-3-ylmethyl)thioformamide, ... | | Authors: | Hanoulle, X, Charton, J, Deprez, B. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Novel dithiocarbamates selectively inhibit 3CL protease of SARS-CoV-2 and other coronaviruses.
Eur.J.Med.Chem., 250, 2023
|
|
1TXK
 
 | | Crystal structure of Escherichia coli OpgG | | Descriptor: | Glucans biosynthesis protein G, SODIUM ION | | Authors: | Hanoulle, X, Rollet, E, Clantin, B, Landrieu, I, Odberg-Ferragut, C, Lippens, G, Bohin, J.P, Villeret, V. | | Deposit date: | 2004-07-05 | | Release date: | 2004-09-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis of Escherichia coli OpgG, a Protein Required for the Biosynthesis of Osmoregulated Periplasmic Glucans.
J.Mol.Biol., 342, 2004
|
|
7P51
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2 MAIN PROTEASE COMPLEXED WITH FRAGMENT F01 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(5-chloropyridin-2-yl)-3-oxo-2,3-dihydro-1H-indene-1-carboxamide, ... | | Authors: | Hanoulle, X, Moschidi, D. | | Deposit date: | 2021-07-13 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
9G13
 
 | | VHH H3-2 in complex with Tau C-terminal peptide | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau, VHH H3-2 | | Authors: | Dupre, E, Landrieu, I, Danis, C, Hanoulle, X, Mortelecque, J. | | Deposit date: | 2024-07-09 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | VHH H3-2 in complex with Tau C-terminal peptide
To Be Published
|
|
5HSV
 
 | | X-Ray structure of a CypA-Alisporivir complex at 1.5 angstrom resolution | | Descriptor: | Alisporivir, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Dujardin, M, Bouckaert, J, Rucktooa, P, Hanoulle, X. | | Deposit date: | 2016-01-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure of alisporivir in complex with cyclophilin A at 1.5 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
2M5L
 
 | |
7QCQ
 
 | | VHH Z70 in interaction with PHF6 Tau peptide | | Descriptor: | GLYCEROL, Tau 301-312, VHH Z70 | | Authors: | Danis, C, Dupre, E, Zejneli, O, Caillierez, R, Arrial, A, Begard, S, Loyens, A, Mortelecque, J, Cantrelle, F.-X, Hanoulle, X, Rain, J.-C, Colin, M, Buee, L, Landrieu, I. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Tau seeding by targeting Tau nucleation core within neurons with a single domain antibody fragment.
Mol.Ther., 30, 2022
|
|
6HT4
 
 | |
8PII
 
 | | VHH Z70 mutant 3 in interaction with PHF6 Tau peptide | | Descriptor: | Microtubule-associated protein tau, VHH Z70 Mutant 3 | | Authors: | Dupre, E, Mortelecque, J, NGuyen, M, Hanoulle, X, Landrieu, I. | | Deposit date: | 2023-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A selection and optimization strategy for single-domain antibodies targeting the PHF6 linear peptide within the tau intrinsically disordered protein.
J.Biol.Chem., 300, 2024
|
|
8OPI
 
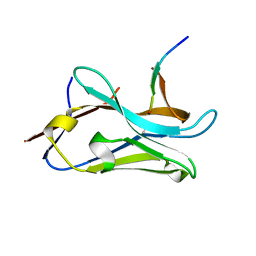 | | VHH Z70 mutant 1 in interaction with PHF6 Tau peptide | | Descriptor: | PHF6 Tau peptide, VHH Z70 mutant 1 | | Authors: | Dupre, E, Mortelecque, J, NGuyen, M, Hanoulle, X, Landrieu, I. | | Deposit date: | 2023-04-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A selection and optimization strategy for single-domain antibodies targeting the PHF6 linear peptide within the tau intrinsically disordered protein.
J.Biol.Chem., 300, 2024
|
|
8OP0
 
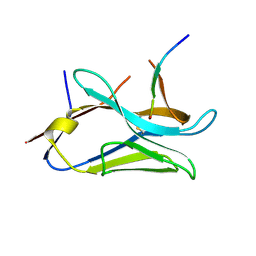 | | VHH Z70 mutant 20 in interaction with PHF6 Tau peptide | | Descriptor: | Tau PHF6 peptide, VHH Z70 mutant 20 | | Authors: | Dupre, E, Mortelecque, J, NGuyen, M, Hanoulle, X, Landrieu, I. | | Deposit date: | 2023-04-06 | | Release date: | 2023-05-31 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A selection and optimization strategy for single-domain antibodies targeting the PHF6 linear peptide within the tau intrinsically disordered protein.
J.Biol.Chem., 300, 2024
|
|
7NLC
 
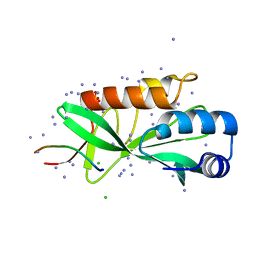 | | Crystallographic structure of human Tsg101 UEV domain in complex with a HEV ORF3 peptide | | Descriptor: | AMMONIUM ION, CHLORIDE ION, Protein ORF3, ... | | Authors: | Moschidi, D, Dupre, E, Villeret, V, Hanoulle, X. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Crystallographic structure of human Tsg101 UEV domain in complex with a HEV ORF3 peptide
To Be Published
|
|
7NTQ
 
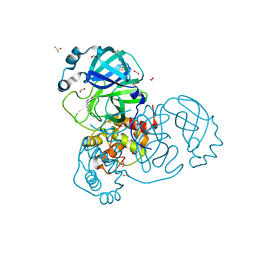 | | Crystal structure of the SARS-CoV-2 Main Protease complexed with N-(pyridin-3-ylmethyl)thioformamide | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Dupre, E, Villeret, V, Hanoulle, X. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Novel dithiocarbamates selectively inhibit 3CL protease of SARS-CoV-2 and other coronaviruses.
Eur.J.Med.Chem., 250, 2023
|
|
7NTT
 
 | |
7NTW
 
 | |
7NTS
 
 | | Crystal structure of the SARS-CoV-2 Main Protease with oxidized C145 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, GLYCEROL, ... | | Authors: | Dupre, E, Villeret, V, Hanoulle, X. | | Deposit date: | 2021-03-10 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.477 Å) | | Cite: | NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5ODD
 
 | | HUMAN MED26 N-TERMINAL DOMAIN (1-92) | | Descriptor: | Mediator of RNA polymerase II transcription subunit 26 | | Authors: | Lens, Z, Cantrelle, F.-X, Perruzini, R, Dewitte, F, Hanoulle, X, Villeret, V, Verger, A, Landrieu, I. | | Deposit date: | 2017-07-05 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N and 13C assignments of the N-terminal domain of the Mediator complex subunit MED26.
Biomol.Nmr Assign., 10, 2016
|
|
