7XN6
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR-deacylization | | 分子名称: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | 著者 | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | 登録日 | 2022-04-28 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
6WQH
 
 | | Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Ig2 substrate, Lon protease, ... | | 著者 | Zhang, K, Li, S, Hsiehb, K, Sub, S, Pintilie, G, Chiu, W, Chang, C. | | 登録日 | 2020-04-28 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Molecular basis for ATPase-powered substrate translocation by the Lon AAA+ protease.
J.Biol.Chem., 297, 2021
|
|
6N34
 
 | | Crystal structure of the BTB domain of Human NS1-BP | | 分子名称: | Influenza virus NS1A-binding protein | | 著者 | Zhang, K, Shang, G, Padavannil, A, Fontoura, B, Chook, Y.M. | | 登録日 | 2018-11-14 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6N3H
 
 | | Crystal structure of Kelch domain of the human NS1 binding protein | | 分子名称: | Influenza virus NS1A-binding protein | | 著者 | Zhang, K, Shang, G, Padavannil, A, Fontoura, B.M.A, Chook, Y.M. | | 登録日 | 2018-11-15 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6NYL
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2c (OA-2c) | | 分子名称: | Vacuolating cytotoxin autotransporter | | 著者 | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | 登録日 | 2019-02-11 | | 公開日 | 2019-03-27 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYN
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2e (OA-2e) | | 分子名称: | Vacuolating cytotoxin autotransporter | | 著者 | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | 登録日 | 2019-02-11 | | 公開日 | 2019-03-27 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8QQ3
 
 | | Streptavidin with a Ni-cofactor | | 分子名称: | 4-[4-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]butylamino]-~{N}1,~{N}1'-di(quinolin-8-yl)cyclohexane-1,1-dicarboxamide, NICKEL (II) ION, Streptavidin | | 著者 | Zhang, K, Jakob, R.P, Ward, T.R. | | 登録日 | 2023-10-03 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | An artificial nickel chlorinase based on the biotin-streptavidin technology.
Chem.Commun.(Camb.), 60, 2024
|
|
7MZ5
 
 | |
7MZ6
 
 | | Cryo-EM structure of minimal TRPV1 with 1 perturbed PI | | 分子名称: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl hexadecanoate, (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, ... | | 著者 | Zhang, K, Julius, D, Cheng, Y. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-22 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZD
 
 | |
6VQX
 
 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF6 | | 分子名称: | AcrF6, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | 著者 | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | 登録日 | 2020-02-06 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UET
 
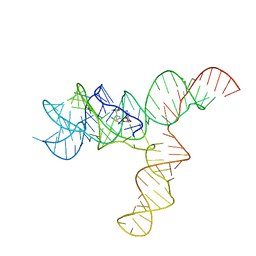 | | SAM-bound SAM-IV riboswitch | | 分子名称: | RNA (119-MER), S-ADENOSYLMETHIONINE | | 著者 | Zhang, K, Li, S, Kappel, K, Pintilie, G, Su, Z, Mou, T, Schmid, M, Das, R, Chiu, W. | | 登録日 | 2019-09-23 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 angstrom resolution.
Nat Commun, 10, 2019
|
|
6UES
 
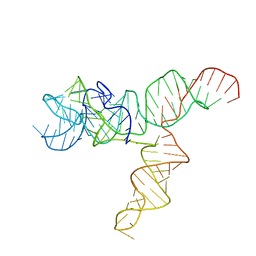 | | Apo SAM-IV Riboswitch | | 分子名称: | RNA (119-MER) | | 著者 | Zhang, K, Li, S, Kappel, K, Pintilie, G, Su, Z, Mou, T, Schmid, M, Das, R, Chiu, W. | | 登録日 | 2019-09-23 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 angstrom resolution.
Nat Commun, 10, 2019
|
|
6XRZ
 
 | | The 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | 分子名称: | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | 著者 | Zhang, K, Zheludev, I, Hagey, R, Wu, M, Haslecker, R, Hou, Y, Kretsch, R, Pintilie, G, Rangan, R, Kladwang, W, Li, S, Pham, E, Souibgui, C, Baric, R, Sheahan, T, Souza, V, Glenn, J, Chiu, W, Das, R. | | 登録日 | 2020-07-14 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Cryo-electron Microscopy and Exploratory Antisense Targeting of the 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome.
Biorxiv, 2020
|
|
5EJO
 
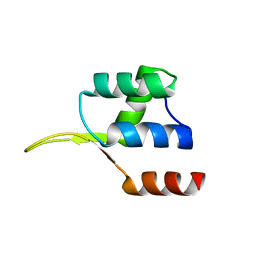 | | Crystal structure of the winged helix domain in Chromatin assembly factor 1 subunit p90 | | 分子名称: | Chromatin assembly factor 1 subunit p90 | | 著者 | Zhang, K, Gao, Y, Li, J, Burgess, R, Han, J, Liang, H, Zhang, Z, Liu, Y. | | 登録日 | 2015-11-02 | | 公開日 | 2016-03-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | A DNA binding winged helix domain in CAF-1 functions with PCNA to stabilize CAF-1 at replication forks
Nucleic Acids Res., 44, 2016
|
|
5FHP
 
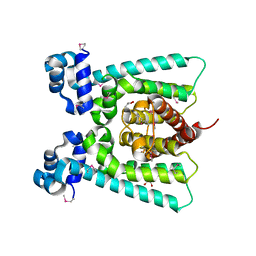 | | SeMet regulator of nicotine degradation | | 分子名称: | GLYCEROL, MALONIC ACID, NicR | | 著者 | Zhang, K, Tang, H, Wu, G, Wang, W, Hu, H, Xu, P. | | 登録日 | 2015-12-22 | | 公開日 | 2016-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Co-crystal Structure of NicR2_Hsp
To Be Published
|
|
7MZB
 
 | | Cryo-EM structure of minimal TRPV1 with 3 bound RTX and 1 perturbed PI | | 分子名称: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | 著者 | Zhang, K, Julius, D, Cheng, Y. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-22 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZC
 
 | |
7MZA
 
 | | Cryo-EM structure of minimal TRPV1 with 2 bound RTX in adjacent pockets | | 分子名称: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | 著者 | Zhang, K, Julius, D, Cheng, Y. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-22 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZ9
 
 | | Cryo-EM structure of minimal TRPV1 with 1 partially bound RTX | | 分子名称: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | 著者 | Zhang, K, Julius, D, Cheng, Y. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-22 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZ7
 
 | |
7MZE
 
 | | Cryo-EM structure of minimal TRPV1 with 2 bound RTX in opposite pockets | | 分子名称: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | 著者 | Zhang, K, Julius, D, Cheng, Y. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-22 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
8CXO
 
 | | Cryo-EM structure of the unliganded mSMO-PGS2 in a lipidic environment | | 分子名称: | CHOLESTEROL, Smoothened homolog, GlgA glycogen synthase chimera | | 著者 | Zhang, K, Wu, H, Hoppe, N, Manglik, A, Cheng, Y. | | 登録日 | 2022-05-22 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Fusion protein strategies for cryo-EM study of G protein-coupled receptors.
Nat Commun, 13, 2022
|
|
5FGL
 
 | | Co-crystal Structure of NicR2_Hsp | | 分子名称: | 4-oxidanylidene-4-(6-oxidanylidene-1~{H}-pyridin-3-yl)butanoic acid, NicR | | 著者 | Zhang, K, Tang, H, Wu, G, Wang, W, Hu, H, Xu, P. | | 登録日 | 2015-12-21 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Co-crystal Structure of NicR2_Hsp
To Be Published
|
|
6VQW
 
 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF8 | | 分子名称: | AcrF8, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | 著者 | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | 登録日 | 2020-02-06 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
