8WYE
 
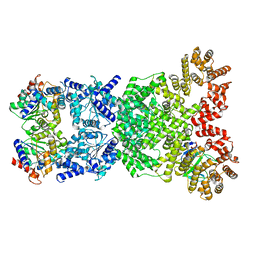 | | Cryo-EM structure of DSR2-DSAD1 (partial) complex | | Descriptor: | Bacillus phage SPbeta DSAD1 protein, SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WY9
 
 | |
8WYA
 
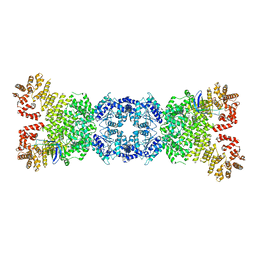 | | Cryo-EM structure of DSR2-tube complex | | Descriptor: | Bacillus phage SPbeta tube protein, SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8XKE
 
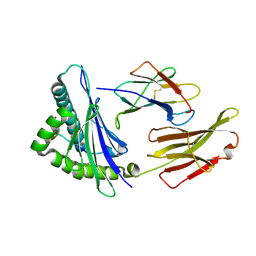 | | The structure of HLA-A/14-3-D | | Descriptor: | Beta-2-microglobulin, GLU-VAL-ASP-ASN-ALA-THR-ARG-PHE-ALA-SER-VAL-TYR, HLA class I heavy chain | | Authors: | Zhang, J.N, Yue, C, Liu, J, Sun, Z.Y. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XFZ
 
 | | The structure of HLA-A/L1-2 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKC
 
 | | The structure of HLA-A/Pep16 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
5ZX5
 
 | | 3.3 angstrom structure of mouse TRPM7 with EDTA | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 7 | | Authors: | Zhang, J, Li, Z, Duan, J, Li, J, Hulse, R.E, Santa-Cruz, A, Abiria, S.A, Krapivinsky, G, Clapham, D.E. | | Deposit date: | 2018-05-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the mammalian TRPM7, a magnesium channel required during embryonic development.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4PXZ
 
 | | Crystal structure of P2Y12 receptor in complex with 2MeSADP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate), CHOLESTEROL, ... | | Authors: | Zhang, J, Zhang, K, Gao, Z.G, Paoletta, S, Zhang, D, Han, G.W, Li, T, Ma, L, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Agonist-bound structure of the human P2Y12 receptor
Nature, 509, 2014
|
|
4GPS
 
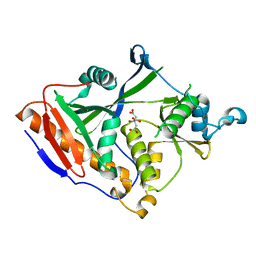 | |
4GPU
 
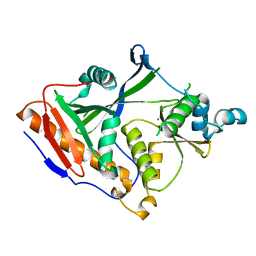 | |
7V5H
 
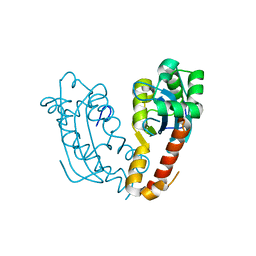 | |
4PY0
 
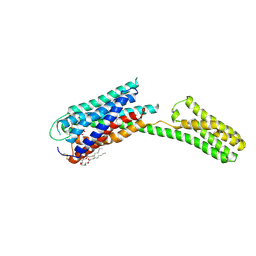 | | Crystal structure of P2Y12 receptor in complex with 2MeSATP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(methylsulfanyl)adenosine 5'-(tetrahydrogen triphosphate), P2Y purinoceptor 12, ... | | Authors: | Zhang, J, Zhang, K, Gao, Z.G, Paoletta, S, Zhang, D, Han, G.W, Li, T, Ma, L, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Agonist-bound structure of the human P2Y12 receptor
Nature, 509, 2014
|
|
8XAI
 
 | |
1M2K
 
 | | Sir2 homologue F159A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2G
 
 | | Sir2 homologue-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2J
 
 | | Sir2 homologue H80N mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2H
 
 | | Sir2 homologue S24A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2N
 
 | |
7V9I
 
 | | The Monomer mutant of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*T)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9H
 
 | | The BEN3 domain of protein Bend3 | | Descriptor: | BEN domain-containing protein 3 | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9F
 
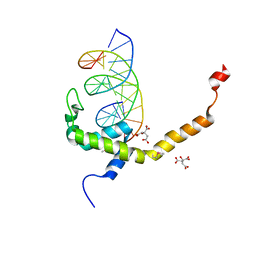 | | Selenomethionine mutant (L740Sem) of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, CITRIC ACID, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), ... | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
3PIF
 
 | |
4J7L
 
 | |
3PIE
 
 | |
4J7M
 
 | |
