7LGH
 
 | |
7LGG
 
 | |
7LGF
 
 | |
7LGE
 
 | |
7LHD
 
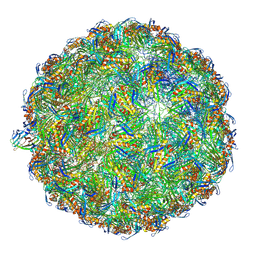 | | The complete model of phage Qbeta virion | | Descriptor: | Capsid protein, Genomic RNA, Maturation protein A2 | | Authors: | Chang, J.Y, Zhang, J. | | Deposit date: | 2021-01-22 | | Release date: | 2022-01-26 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Assembly of Q beta Virion and Its Diverse Forms of Virus-like Particles.
Viruses, 14, 2022
|
|
7Y4F
 
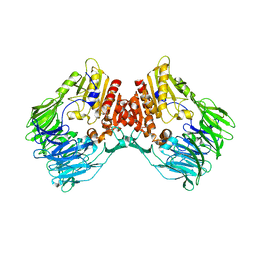 | | bacterial DPP4 | | Descriptor: | Dipeptidyl peptidase IV | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
7Y4G
 
 | | sit-bound btDPP4 | | Descriptor: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, btDPP4 | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
8HAY
 
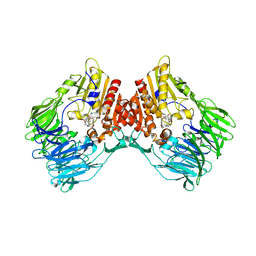 | | d4-bound btDPP4 | | Descriptor: | (1~{R})-1-[[4-[5-[[(1~{R})-6,7-dimethoxy-2-methyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]methyl]-2-methoxy-phenoxy]phenyl]methyl]-6,7-dimethoxy-2-methyl-3,4-dihydro-1~{H}-isoquinoline, btDPP4 | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-10-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
9CBS
 
 | |
8D56
 
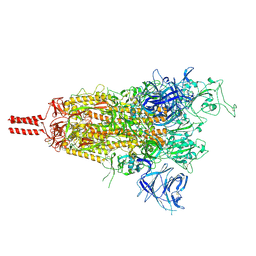 | | One RBD-up state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8D55
 
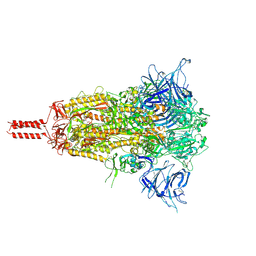 | | Closed state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8D5A
 
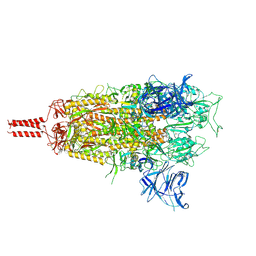 | | Middle state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8J6M
 
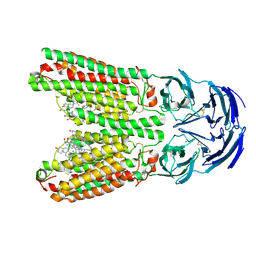 | | SIDT1 protein | | Descriptor: | CHOLESTEROL, Green fluorescent protein,SID1 transmembrane family member 1, OLEIC ACID, ... | | Authors: | Zhang, J.T, Jiang, D.H. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8ISM
 
 | | HSA-Pt compound complex | | Descriptor: | 7-(2-azanyl-5-chloranyl-phenyl)-3$l^{3}-thia-5,6$l^{4}-diaza-2$l^{3}-platinatricyclo[6.4.0.0^{2,6}]dodeca-1(12),3,6,8,10-pentaen-4-amine, PALMITIC ACID, Serum albumin | | Authors: | Zhang, J.Z, Zhang, Z.L. | | Deposit date: | 2023-03-21 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | HSA-Pt compound complex
To Be Published
|
|
7XM9
 
 | | Cryo-EM structure of human NaV1.7/beta1/beta2-XEN907 | | Descriptor: | (7~{R})-1'-pentylspiro[6~{H}-furo[3,2-f][1,3]benzodioxole-7,3'-indole]-2'-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | zhang, J.T, Jiang, D.H. | | Deposit date: | 2022-04-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XMF
 
 | | Cryo-EM structure of human NaV1.7/beta1/beta2-Nav1.7-IN2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[[4-[3-(4-fluoranyl-2-methyl-phenoxy)azetidin-1-yl]pyrimidin-2-yl]amino]-~{N}-methyl-benzamide, ... | | Authors: | Zhang, J.T, Jiang, D.H. | | Deposit date: | 2022-04-25 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XR5
 
 | | Crystal structure of imine reductase with NAPDH from Streptomyces albidoflavus | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45,48,51,54,57-nonadecaoxanonapentacontane-1,59-diol, 6-phosphogluconate dehydrogenase NAD-binding, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J, Chen, R.C, Gao, S.S. | | Deposit date: | 2022-05-09 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Actinomycetes-derived imine reductases with a preference towards bulky amine substrates.
Commun Chem, 5, 2022
|
|
8Y82
 
 | | Cryo-EM structure of the tetrameric SPARSA gRNA-ssDNA-NAD+ complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (25-mer), MAGNESIUM ION, ... | | Authors: | Zhang, J.T, Cui, N, Wei, X.Y, Jia, N. | | Deposit date: | 2024-02-05 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Tetramerization-dependent activation of the Sir2-associated short prokaryotic Argonaute immune system.
Nat Commun, 15, 2024
|
|
8Y80
 
 | | Cryo-EM structure of the tetrameric SPARSA gRNA-ssDNA complex | | Descriptor: | DNA (25-mer), MAGNESIUM ION, Piwi domain protein, ... | | Authors: | Zhang, J.T, Cui, N, Wei, X.Y, Jia, N. | | Deposit date: | 2024-02-05 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Tetramerization-dependent activation of the Sir2-associated short prokaryotic Argonaute immune system.
Nat Commun, 15, 2024
|
|
7XE8
 
 | |
8Y7Z
 
 | | Cryo-EM structure of the monomeric SPARSA gRNA-ssDNA complex | | Descriptor: | DNA (25-mer), MAGNESIUM ION, Piwi domain protein, ... | | Authors: | Zhang, J.T, Cui, N, Wei, X.Y, Jia, N. | | Deposit date: | 2024-02-05 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Tetramerization-dependent activation of the Sir2-associated short prokaryotic Argonaute immune system.
Nat Commun, 15, 2024
|
|
7XG0
 
 | | CryoEM structure of type IV-A Csf-crRNA-dsDNA ternary complex | | Descriptor: | Csf1, Csf2, Csf3, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-04-02 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Type IV-A CRISPR-Csf complex: Assembly, dsDNA targeting, and CasDinG recruitment.
Mol.Cell, 83, 2023
|
|
7XG4
 
 | | CryoEM structure of type IV-A CasDinG bound NTS-nicked Csf-crRNA-dsDNA quaternary complex in a second state | | Descriptor: | Csf1, Csf2, Csf3, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-04-02 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Type IV-A CRISPR-Csf complex: Assembly, dsDNA targeting, and CasDinG recruitment.
Mol.Cell, 83, 2023
|
|
7XG2
 
 | | CryoEM structure of type IV-A NTS-nicked dsDNA bound Csf-crRNA ternary complex | | Descriptor: | Csf1, Csf2, Csf3, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-04-02 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Type IV-A CRISPR-Csf complex: Assembly, dsDNA targeting, and CasDinG recruitment.
Mol.Cell, 83, 2023
|
|
7XG3
 
 | | CryoEM structure of type IV-A CasDinG bound NTS-nicked Csf-crRNA-dsDNA quaternary complex | | Descriptor: | Csf1, Csf2, Csf3, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-04-02 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Type IV-A CRISPR-Csf complex: Assembly, dsDNA targeting, and CasDinG recruitment.
Mol.Cell, 83, 2023
|
|
