7FAD
 
 | |
5BN4
 
 | | Structure of a unique ATP synthase NeqA-NeqB in complex with ANP from Nanoarcheaum equitans | | Descriptor: | MAGNESIUM ION, NEQ263, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
5KW9
 
 | |
5BO5
 
 | | Structure of a unique ATP synthase subunit NeqB from Nanoarcheaum equitans | | Descriptor: | MAGNESIUM ION, NEQ263, SULFATE ION | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
4QTK
 
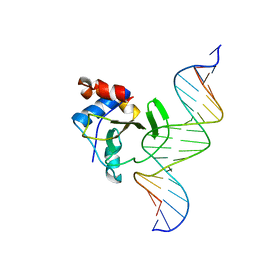 | | Complex of WOPR domain of Wor1 in Candida albicans with the 17bp dsDNA | | Descriptor: | DNA (5'-D(*AP*AP*GP*TP*TP*AP*AP*AP*CP*TP*TP*TP*TP*TP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*AP*AP*AP*AP*AP*GP*TP*TP*TP*AP*AP*CP*TP*T)-3'), White-opaque regulator 1 | | Authors: | Zhang, S, Zhang, T, Ding, J. | | Deposit date: | 2014-07-08 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of the WOPR-DNA complex and implications for Wor1 function in white-opaque switching of Candida albicans.
Cell Res., 24, 2014
|
|
3H16
 
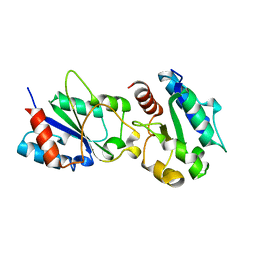 | | Crystal structure of a bacteria TIR domain, PdTIR from Paracoccus denitrificans | | Descriptor: | SULFATE ION, TIR protein | | Authors: | Chan, S.L, Low, L.Y, Santelli, E, Pascual, J. | | Deposit date: | 2009-04-11 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mimicry in Innate Immunity: CRYSTAL STRUCTURE OF A BACTERIAL TIR DOMAIN.
J.Biol.Chem., 284, 2009
|
|
2JRK
 
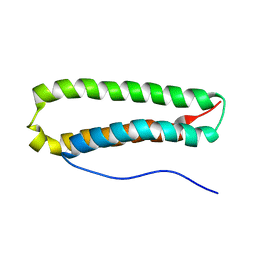 | | NMR Structure and Epitope Mapping of Blo t 5 | | Descriptor: | Mite allergen Blo t 5 | | Authors: | Chan, S, Mok, Y. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure and IgE epitopes of Blo t 5, a major dust mite allergen
J.Immunol., 181, 2008
|
|
7WB0
 
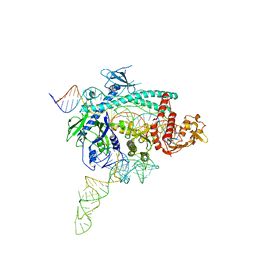 | |
7WAZ
 
 | |
7WAY
 
 | |
7WB1
 
 | |
3IR2
 
 | |
2BYP
 
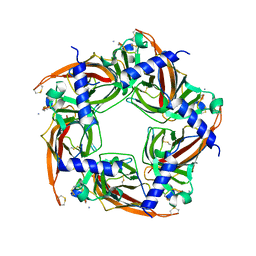 | | Crystal structure of Aplysia californica AChBP in complex with alpha- conotoxin ImI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-CONOTOXIN IMI, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Hansen, S.B, Sulzenbacher, G, Huxford, T, Marchot, P, Taylor, P, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structures of Aplysia Achbp Complexes with Nicotinic Agonists and Antagonists Reveal Distinctive Binding Interfaces and Conformations.
Embo J., 24, 2005
|
|
3A59
 
 | |
2BV3
 
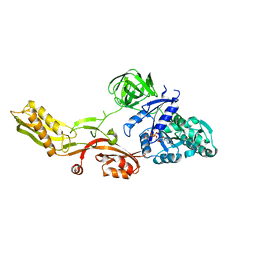 | | Crystal structure of a mutant elongation factor G trapped with a GTP analogue | | Descriptor: | ELONGATION FACTOR G, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Hansson, S, Singh, R, Gudkov, A.T, Liljas, A, Logan, D.T. | | Deposit date: | 2005-06-22 | | Release date: | 2005-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Mutant Elongation Factor G Trapped with a GTP Analogue.
FEBS Lett., 579, 2005
|
|
3IRK
 
 | | Solution Structure of Heparin dp30 | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose | | Authors: | Khan, S, Gor, J, Mulloy, B, Perkins, S.J. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: new insight into heparin-protein complexes.
J.Mol.Biol., 395, 2010
|
|
4LF1
 
 | | Hexameric Form II RuBisCO from Rhodopseudomonas palustris, activated and complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Chan, S, Satagopan, S, Sawaya, M.R, Eisenberg, D, Tabita, F.R, Perry, L.J. | | Deposit date: | 2013-06-26 | | Release date: | 2014-06-25 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure-function studies with the unique hexameric form II ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Rhodopseudomonas palustris.
J.Biol.Chem., 289, 2014
|
|
7W3H
 
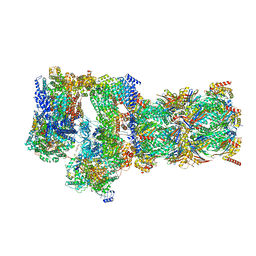 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED2.1_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W39
 
 | | Structure of USP14-bound human 26S proteasome in state EA2.1_UBL | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3B
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED5_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3G
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED2.0_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3J
 
 | | Structure of USP14-bound human 26S proteasome in substrate-inhibited state SC_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3A
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED4_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3C
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED0_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W37
 
 | | Structure of USP14-bound human 26S proteasome in state EA1_UBL | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
