3D2T
 
 | |
1ZUG
 
 | | STRUCTURE OF PHAGE 434 CRO PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | PHAGE 434 CRO PROTEIN | | Authors: | Padmanabhan, S, Jimenez, M.A, Gonzalez, C, Sanz, J.M, Gimenez-Gallego, G, Rico, M. | | Deposit date: | 1997-03-14 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and stability of phage 434 Cro protein.
Biochemistry, 36, 1997
|
|
1Y67
 
 | |
5OWF
 
 | | Structure of a LAO-binding protein mutant with glutamine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLUTAMINE, ... | | Authors: | Shanmugaratnam, S, Banda-Vazquez, J, Sosa-Peinado, A, Hocker, B. | | Deposit date: | 2017-08-31 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Redesign of LAOBP to bind novel l-amino acid ligands.
Protein Sci., 27, 2018
|
|
3Q3Q
 
 | |
4H02
 
 | |
1R9X
 
 | | Bacterial cytosine deaminase D314G mutant. | | Descriptor: | Cytosine deaminase, FE (III) ION, GLYCEROL, ... | | Authors: | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
2KE8
 
 | | NMR solution structure of metal-modified DNA | | Descriptor: | DNA (5'-D(*TP*TP*AP*AP*TP*TP*TP*(D33)P*(D33)P*(D33)P*AP*AP*AP*TP*TP*AP*A)-3'), SILVER ION | | Authors: | Johannsen, S, Duepre, N, Boehme, D, Mueller, J, Sigel, R.K.O. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA double helix with consecutive metal-mediated base pairs.
Nat.Chem., 2, 2010
|
|
2K67
 
 | |
4JFA
 
 | | Crystal Structure of Plasmodium falciparum Tryptophanyl-tRNA synthetase | | Descriptor: | BETA-MERCAPTOETHANOL, POTASSIUM ION, TRYPTOPHAN, ... | | Authors: | Khan, S, Garg, A, Manickam, Y, Sharma, A. | | Deposit date: | 2013-02-28 | | Release date: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An appended domain results in an unusual architecture for malaria parasite tryptophanyl-tRNA synthetase
Plos One, 8, 2013
|
|
4R42
 
 | | Crystal structure of KatB, a manganese catalase from Anabaena PCC7120 | | Descriptor: | Alr3090 protein, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bihani, S.C. | | Deposit date: | 2014-08-19 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | KatB, a cyanobacterial Mn-catalase with unique active site configuration: Implications for enzyme function.
Free Radic. Biol. Med., 93, 2016
|
|
1Z17
 
 | | Crystal structure analysis of periplasmic Leu/Ile/Val-binding protein with bound ligand isoleucine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ISOLEUCINE, Leu/Ile/Val-binding protein | | Authors: | Trakhanov, S.D, Vyas, N.K, Kristensen, D.M, Ma, J, Quiocho, F.A. | | Deposit date: | 2005-03-03 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity.
Biochemistry, 44, 2005
|
|
1Z16
 
 | | Crystal structure analysis of periplasmic Leu/Ile/Val-binding protein with bound leucine | | Descriptor: | CADMIUM ION, LEUCINE, Leu/Ile/Val-binding protein | | Authors: | Trakhanov, S.D, Vyas, N.K, Kristensen, D.M, Ma, J, Quiocho, F.A. | | Deposit date: | 2005-03-03 | | Release date: | 2005-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity.
Biochemistry, 44, 2005
|
|
1QF4
 
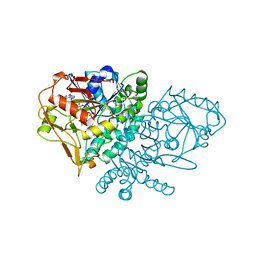 | | DESIGN, SYNTHESIS, AND X-RAY CRYSTAL STRUCTURE OF AN ENZYME BOUND BISUBSTRATE HYBRID INHIBITOR OF ADENYLOSUCCINATE SYNTHETASE | | Descriptor: | (C8-R)-HYDANTOCIDIN 5'-PHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hanessian, S, Lu, P.-P, Sanceau, J.-Y, Chemla, P, Gohda, K, Fonne-Pfister, R, Prade, L, Cowan-Jacob, S.W. | | Deposit date: | 1999-04-06 | | Release date: | 1999-12-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An enzyme-bound bisubstrate hybrid inhibitor of adenylosuccinate synthetase
Angew.Chem.Int.Ed.Engl., 38, 1999
|
|
1Z8F
 
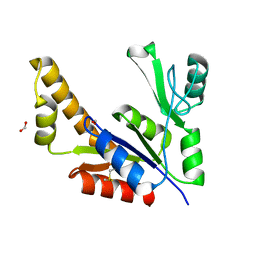 | | Guanylate Kinase Double Mutant A58C, T157C from Mycobacterium tuberculosis (Rv1389) | | Descriptor: | FORMIC ACID, Guanylate kinase | | Authors: | Chan, S, Sawaya, M.R, Choi, B, Zocchi, G, Perry, L.J, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2005-03-30 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Guanylate Kinase from Mycobacterium tuberculosis (Rv1389)
To be Published
|
|
1Z18
 
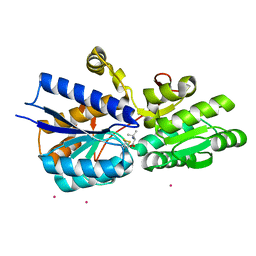 | | Crystal structure analysis of periplasmic Leu/Ile/Val-binding protein with bound valine | | Descriptor: | CADMIUM ION, Leu/Ile/Val-binding protein, VALINE | | Authors: | Trakhanov, S.D, Vyas, N.K, Kristensen, D.M, Ma, J, Quiocho, F.A. | | Deposit date: | 2005-03-03 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity.
Biochemistry, 44, 2005
|
|
1Z15
 
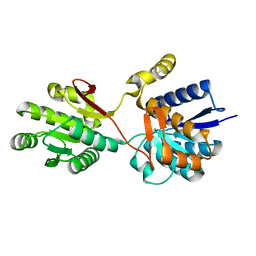 | | Crystal structure analysis of periplasmic Leu/Ile/Val-binding protein in superopen form | | Descriptor: | Leu/Ile/Val-binding protein | | Authors: | Trakhanov, S.D, Vyas, N.K, Kristensen, D.M, Ma, J, Quiocho, F.A. | | Deposit date: | 2005-03-03 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity.
Biochemistry, 44, 2005
|
|
1QF5
 
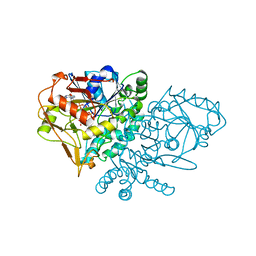 | | DESIGN, SYNTHESIS, AND X-RAY CRYSTAL STRUCTURE OF AN ENZYME BOUND BISUBSTRATE HYBRID INHIBITOR OF ADENYLOSUCCINATE SYNTHETASE | | Descriptor: | (C8-S)-HYDANTOCIDIN 5'-PHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hanessian, S, Lu, P.-P, Sanceau, J.-Y, Chemla, P, Prade, L, Gohda, K, Cowan-Jacob, S.W, Fonne-Pfister, R. | | Deposit date: | 1999-04-06 | | Release date: | 1999-12-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An enzyme-bound bisubstrate hybrid inhibitor of adenylosuccinate synthetase
Angew.Chem.Int.Ed.Engl., 38, 1999
|
|
1QWV
 
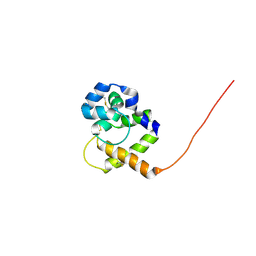 | |
3PYD
 
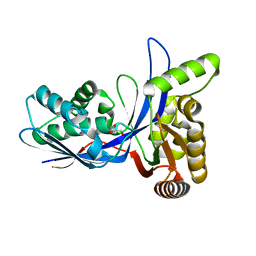 | | crystal structure of 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (IspE) from Mycobacterium tuberculosis | | Descriptor: | 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase, GLYCEROL | | Authors: | Shan, S, Chen, X.H, Liu, T, Zhao, H.C, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2010-12-13 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | The Structural Basis for anti-TB Drug Discovery targeting of 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (IspE) from Mycobacterium tuberculosis
To be Published
|
|
4P3I
 
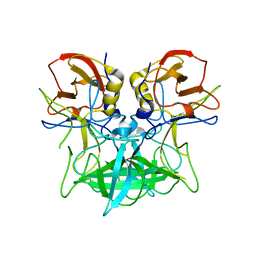 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeA HBGA. | | Descriptor: | P domain of VP1, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6941 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
1EPW
 
 | |
4P12
 
 | | Native Structure of the P domain from a GI.7 Norovirus variant. | | Descriptor: | Major capsid protein | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-02-24 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6011 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P25
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeY HBGA. | | Descriptor: | Major capsid protein, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4991 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
2K45
 
 | |
