3S6J
 
 | | The crystal structure of a hydrolase from Pseudomonas syringae | | Descriptor: | CALCIUM ION, Hydrolase, haloacid dehalogenase-like family | | Authors: | Zhang, Z, Syed Ibrahim, B, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-05-25 | | Release date: | 2011-07-13 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | The crystal structure of a hydrolase from Pseudomonas syringae
To be Published
|
|
3RHE
 
 | |
3R03
 
 | |
4DIO
 
 | | The crystal structure of transhydrogenase from Sinorhizobium meliloti | | Descriptor: | NAD(P) transhydrogenase subunit alpha part 1 | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-01-31 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of transhydrogenase from Sinorhizobium meliloti
To be Published
|
|
3ZN8
 
 | | Structural Basis of Signal Sequence Surveillance and Selection by the SRP-SR Complex | | Descriptor: | 4.5 S RNA, DIPEPTIDYL AMINOPEPTIDASE B, MAGNESIUM ION, ... | | Authors: | von Loeffelholz, O, Knoops, K, Ariosa, A, Zhang, X, Karuppasamy, M, Huard, K, Schoehn, G, Berger, I, Shan, S.O, Schaffitzel, C. | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural Basis of Signal Sequence Surveillance and Selection by the Srp-Sr Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
4DRY
 
 | | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase from Rhizobium meliloti | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, SULFATE ION | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-17 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase from Rhizobium meliloti
To be Published
|
|
3SSZ
 
 | | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Rhodobacteraceae bacterium | | Descriptor: | Mandelate racemase/muconate lactonizing enzyme, N-terminal domain protein, SULFATE ION | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Rhodobacteraceae bacterium
To be Published
|
|
3LDY
 
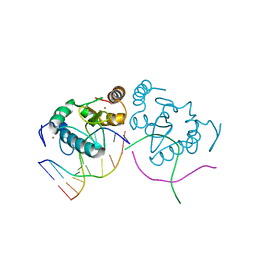 | | An extraordinary mechanism of DNA perturbation exhibited by the rare-cutting HNH restriction endonuclease PacI | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*AP*GP*GP*CP*TP*TP*A)-3'), DNA (5'-D(P*AP*TP*TP*AP*AP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Shen, B.W, Heiter, D, Chan, S.-H, Xu, S.-Y, Wilson, G, Stoddard, B.L. | | Deposit date: | 2010-01-13 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI.
Structure, 18, 2010
|
|
8JJ9
 
 | |
3LQ7
 
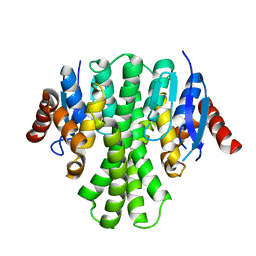 | | Crystal structure of glutathione s-transferase from agrobacterium tumefaciens str. c58 | | Descriptor: | Glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-08 | | Release date: | 2010-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Agrobacterium Tumefaciens
To be Published
|
|
8J66
 
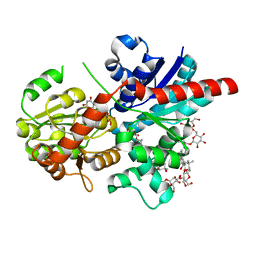 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M3, state 2 | | Descriptor: | (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2R,3S,4S,5R,6S)-6-[(3R,6S)-6-[(3S,8S,9R,10R,11S,13R,14S,17S)-3-[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-2-oxidanyl-heptan-3-yl]oxy-3,4,5-tris(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2023-04-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
6P3Q
 
 | | Calpain-5 (CAPN5) Protease Core (PC) | | Descriptor: | Calpain-5 | | Authors: | Velez, G, Sun, Y.J, Khan, S, Yang, J, Koster, H.J, Lokesh, G, Mahajan, V. | | Deposit date: | 2019-05-24 | | Release date: | 2020-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Unique Activation Mechanisms of a Non-classical Calpain and Its Disease-Causing Variants.
Cell Rep, 30, 2020
|
|
8AT0
 
 | | PotF with mutation D247K | | Descriptor: | (2R)-1-methoxypropan-2-amine, (2~{S})-1-(2-methoxyethoxy)propan-2-amine, 1,2-ETHANEDIOL, ... | | Authors: | Kroeger, P, Shanmugaratnam, S, Hocker, B. | | Deposit date: | 2022-08-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A fluorescent biosensor for the visualization of Agmatine
To Be Published
|
|
6RVU
 
 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) | | Descriptor: | 1,2-ETHANEDIOL, Lethal Factor 1 (BLF1) | | Authors: | Mobbs, G.W, Aziz, A.A, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2019-06-01 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
6LZ0
 
 | | Cryo-EM structure of human MCT1 in complex with Basigin-2 in the presence of lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
3LSZ
 
 | | Crystal structure of glutathione s-transferase from Rhodobacter sphaeroides | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTATHIONE, GLYCEROL, ... | | Authors: | Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-14 | | Release date: | 2010-03-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glutathione s-transferase from Rhodobacter sphaeroides
To be Published
|
|
5M7U
 
 | | Structure of human O-GlcNAc hydrolase with new iminocyclitol type inhibitor | | Descriptor: | 2-[(2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-[3-[3-(trifluoromethyl)phenyl]propyl]pyrrolidin-2-yl]-~{N}-methyl-ethanamide, Protein O-GlcNAcase | | Authors: | Roth, C, Chan, S, Offen, W.A, Hemsworth, G.R, Willems, L.I, King, D, Varghese, V, Britton, R, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional insight into human O-GlcNAcase.
Nat. Chem. Biol., 13, 2017
|
|
3LXT
 
 | |
4F7T
 
 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009 H1N1 PB1 (498-505) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
4RIS
 
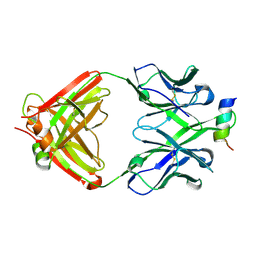 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | Descriptor: | CH58-UA Fab heavy chain, CH58-UA Fab light chain, Envelope glycoprotein | | Authors: | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
4RIR
 
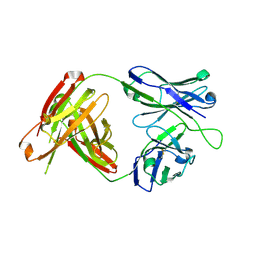 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | Descriptor: | CH58-UA Fab heavy chain, CH58-UA Fab light chain | | Authors: | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
5NCO
 
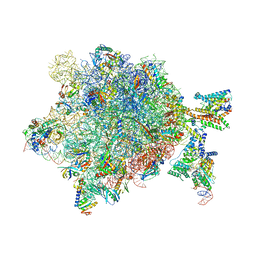 | | Quaternary complex between SRP, SR, and SecYEG bound to the translating ribosome | | Descriptor: | 23S rRNA, 4.5S SRP RNA (Ffs), 50S ribosomal protein L10, ... | | Authors: | Jomaa, A, Hwang Fu, Y, Boerhinger, D, Leibundgut, M, Shan, S.O, Ban, N. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2018-03-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome.
Nat Commun, 8, 2017
|
|
4F7M
 
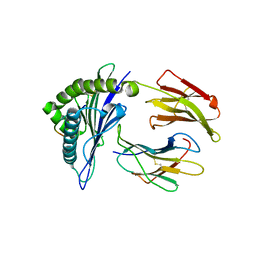 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009 H1N1 PA (649-658) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, George, F.G. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
6Q84
 
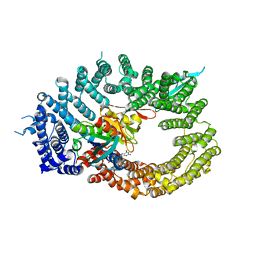 | | Crystal structure of RanGTP-Pdr6-eIF5A export complex | | Descriptor: | Eukaryotic translation initiation factor 5A-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Aksu, M, Trakhanov, S, Vera-Rodriguez, A, Gorlich, D. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122.
J.Cell Biol., 218, 2019
|
|
3MKC
 
 | |
