6PNJ
 
 | | Structure of Photosystem I Acclimated to Far-red Light | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | 著者 | Gisriel, C.J, Shen, G, Kurashov, V, Ho, M, Zhang, S, Williams, D, Golbeck, J.H, Fromme, P, Bryant, D.A. | | 登録日 | 2019-07-02 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-02-26 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The structure of Photosystem I acclimated to far-red light illuminates an ecologically important acclimation process in photosynthesis
Sci Adv, 6, 2020
|
|
2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | 分子名称: | hypothetical protein | | 著者 | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2006-02-09 | | 公開日 | 2006-02-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
6M20
 
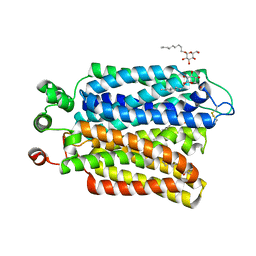 | | Crystal structure of Plasmodium falciparum hexose transporter PfHT1 bound with glucose | | 分子名称: | Hexose transporter 1, beta-D-glucopyranose, nonyl beta-D-glucopyranoside | | 著者 | Jiang, X, Yuan, Y.Y, Zhang, S, Wang, N, Yan, C.Y, Yan, N. | | 登録日 | 2020-02-26 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Basis for Blocking Sugar Uptake into the Malaria Parasite Plasmodium falciparum.
Cell, 183, 2020
|
|
5W96
 
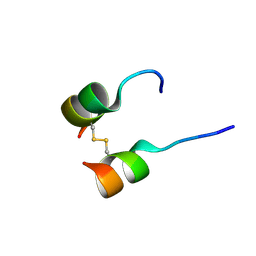 | | Solution structure of phage derived peptide inhibitor of frizzled 7 receptor | | 分子名称: | Fz7 binding peptide | | 著者 | Nile, A.H, de Sousa e Melo, F, Mukund, S, Piskol, R, Hansen, S, Zhou, L, Zhang, Y, Fu, Y, Gogol, E.B, Komuves, L.G, Modrusan, Z, Angers, S, Franke, Y, Koth, C, Fairbrother, W.J, Wang, W, de Sauvage, F.J, Hannoush, R.N. | | 登録日 | 2017-06-22 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
1OZO
 
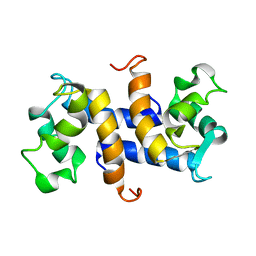 | | Three-dimensional solution structure of apo-S100P protein determined by NMR spectroscopy | | 分子名称: | S-100P protein | | 著者 | Lee, Y.-C, Volk, D.E, Thiviyanathan, V, Kleerekoper, Q, Gribenko, A.V, Zhang, S, Gorenstein, D.G, Makhatadze, G.I, Luxon, B.A. | | 登録日 | 2003-04-09 | | 公開日 | 2004-04-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the Apo-S100P protein.
J.Biomol.Nmr, 29, 2004
|
|
3HH0
 
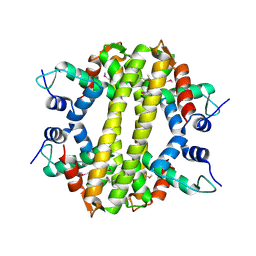 | | Crystal structure of a transcriptional regulator, MerR family from Bacillus cereus | | 分子名称: | Transcriptional regulator, MerR family | | 著者 | Palani, K, Zhang, Z, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-05-14 | | 公開日 | 2009-05-26 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Crystal structure of a transcriptional regulator, MerR family from Bacillus cereus
To be Published
|
|
5J87
 
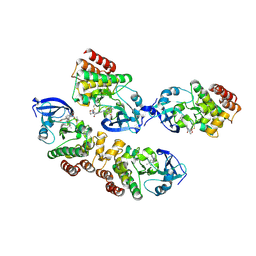 | |
7PQ0
 
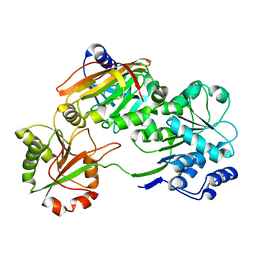 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) C94S inactive mutant in complex with human eIF4A - Crystal form B | | 分子名称: | Burkholderia Lethal Factor 1 (BLF1), Eukaryotic initiation factor 4A-I | | 著者 | Mobbs, G.W, Aziz, A.A, Dix, S.R, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | 登録日 | 2021-09-15 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
7PPZ
 
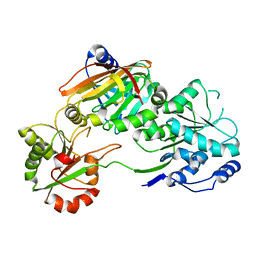 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) C94S inactive mutant in complex with human eIF4A - Crystal form A | | 分子名称: | Burkholderia Lethal Factor 1 (BLF1), Eukaryotic initiation factor 4A-I | | 著者 | Mobbs, G.W, Aziz, A.A, Dix, S.R, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | 登録日 | 2021-09-15 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
7PZT
 
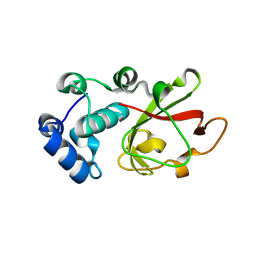 | | Structure of the bacterial toxin, TecA, an asparagine deamidase from Alcaligenes faecalis. | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Urea amidohydrolase | | 著者 | Dix, S.R, Aziz, A.A, Baker, P.J, Evans, C.A, Dickman, M.J, Farthing, R.J, King, Z.L.S, Nathan, S, Partridge, L.J, Raih, F.M, Sedelnikova, S.E, Thomas, M.S, Rice, D.W. | | 登録日 | 2021-10-13 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | The structure of A. faecalis TecA provides insights into its role as an asparagine deamidase toxin which targets RhoA
To Be Published
|
|
7BPI
 
 | | The crystal structue of PDE10A complexed with 14 | | 分子名称: | 8-[(E)-2-[5-methyl-1-[3-[3-(4-methylpiperazin-1-yl)propoxy]phenyl]benzimidazol-2-yl]ethenyl]quinoline, MAGNESIUM ION, ZINC ION, ... | | 著者 | Yang, Y, Zhang, S, Zhou, Q, Huang, Y.-Y, Guo, L, Luo, H.-B. | | 登録日 | 2020-03-22 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4000864 Å) | | 主引用文献 | Discovery of highly selective and orally available benzimidazole-based phosphodiesterase 10 inhibitors with improved solubility and pharmacokinetic properties for treatment of pulmonary arterial hypertension.
Acta Pharm Sin B, 10, 2020
|
|
8P1U
 
 | | Structure of divisome complex FtsWIQLB | | 分子名称: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ, ... | | 著者 | Yang, L, Chang, S, Tang, D, Dong, H. | | 登録日 | 2023-05-12 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural insights into the activation of the divisome complex FtsWIQLB.
Cell Discov, 10, 2024
|
|
7AOP
 
 | | Structure of NUDT15 in complex with inhibitor TH8321 | | 分子名称: | 2-azanyl-9-cyclohexyl-8-(2-methoxyphenyl)-3~{H}-purine-6-thione, MAGNESIUM ION, Nucleotide triphosphate diphosphatase NUDT15 | | 著者 | Rehling, D, Zhang, S.M, Helleday, T, Stenmark, P. | | 登録日 | 2020-10-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | NUDT15-mediated hydrolysis limits the efficacy of anti-HCMV drug ganciclovir.
Cell Chem Biol, 28, 2021
|
|
7AOM
 
 | | Structure of NUDT15 in complex with Ganciclovir triphosphate | | 分子名称: | Ganciclovir triphosphate, MAGNESIUM ION, Nucleotide triphosphate diphosphatase NUDT15 | | 著者 | Rehling, D, Zhang, S.M, Helleday, T, Stenmark, P. | | 登録日 | 2020-10-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | NUDT15-mediated hydrolysis limits the efficacy of anti-HCMV drug ganciclovir.
Cell Chem Biol, 28, 2021
|
|
1ML9
 
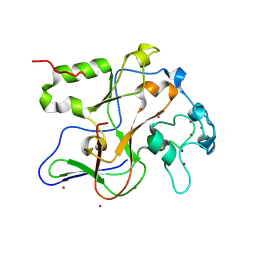 | | Structure of the Neurospora SET domain protein DIM-5, a histone lysine methyltransferase | | 分子名称: | Histone H3 methyltransferase DIM-5, UNKNOWN, ZINC ION | | 著者 | Zhang, X, Tamaru, H, Khan, S.I, Horton, J.R, Keefe, L.J, Selker, E.U, Cheng, X. | | 登録日 | 2002-08-30 | | 公開日 | 2002-10-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structure of the Neurospora SET domain protein DIM-5,
a histone H3 lysine methyltransferase
Cell(Cambridge,Mass.), 111, 2002
|
|
3TU8
 
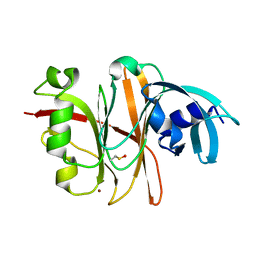 | | Crystal Structure of the Burkholderia Lethal Factor 1 (BLF1) | | 分子名称: | BROMIDE ION, Burkholderia Lethal Factor 1 (BLF1) | | 著者 | Cruz, A, Hautbergue, G.M, Artymiuk, P.J, Baker, P.J, Chang, C.T, Mahadi, N.M, Mobbs, G.W, Mohamed, R, Nathan, S, Partridge, L.J, Raih, M.F, Ruzheinikov, S.N, Sedelnikova, S.E, Wilson, S.A, Rice, D.W. | | 登録日 | 2011-09-16 | | 公開日 | 2011-11-30 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | A Burkholderia pseudomallei toxin inhibits helicase activity of translation factor eIF4A.
Science, 334, 2011
|
|
7RHA
 
 | | A new fluorescent protein darkmRuby at pH 5.0 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | 著者 | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | 登録日 | 2021-07-16 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a new fluorescent protein darkmRuby at pH 5.0
To Be Published
|
|
7OXL
 
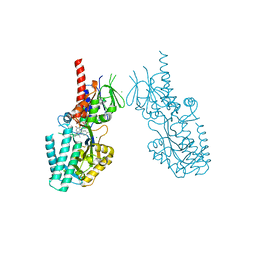 | | Crystal structure of human Spermine Oxidase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FAD-MDL72527 adduct, ... | | 著者 | Impagliazzo, A, Johannsson, S, Thomsen, M, Krapp, S. | | 登録日 | 2021-06-22 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-08-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of human spermine oxidase in complex with a highly selective allosteric inhibitor.
Commun Biol, 5, 2022
|
|
7OY0
 
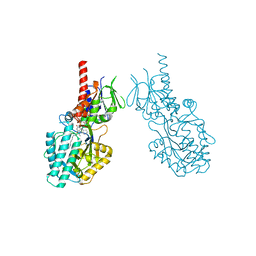 | | Structure of human Spermine Oxidase in complex with a highly selective allosteric inhibitor | | 分子名称: | 4-[(4-imidazo[1,2-a]pyridin-3-yl-1,3-thiazol-2-yl)amino]phenol, CHLORIDE ION, FAD-MDL72527 adduct, ... | | 著者 | Impagliazzo, A, Thomsen, M, Johannsson, S, Krapp, S. | | 登録日 | 2021-06-23 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-08-17 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structure of human spermine oxidase in complex with a highly selective allosteric inhibitor.
Commun Biol, 5, 2022
|
|
6LZ0
 
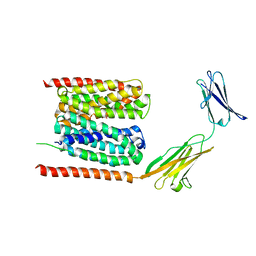 | | Cryo-EM structure of human MCT1 in complex with Basigin-2 in the presence of lactate | | 分子名称: | (2S)-2-HYDROXYPROPANOIC ACID, Basigin, Monocarboxylate transporter 1 | | 著者 | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | 登録日 | 2020-02-16 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
3TUA
 
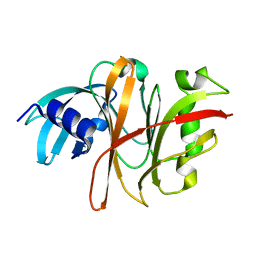 | | Crystal Structure of the Burkholderia Lethal Factor 1 (BLF1) C94S mutant | | 分子名称: | Burkholderia Lethal Factor 1 (BLF1) | | 著者 | Cruz, A, Hautbergue, G.M, Artymiuk, P.J, Baker, P.J, Chang, C.T, Mahadi, N.M, Mobbs, G.W, Mohamed, R, Nathan, S, Partridge, L.J, Raih, M.F, Ruzheinikov, S.N, Sedelnikova, S.E, Wilson, S.A, Rice, D.W. | | 登録日 | 2011-09-16 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | A Burkholderia pseudomallei toxin inhibits helicase activity of translation factor eIF4A.
Science, 334, 2011
|
|
6MAU
 
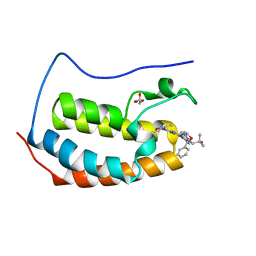 | | Crystal structure of human BRD4(1) in complex with CN210 (compound 19) | | 分子名称: | 1-(4-{6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-[(3S)-3-phenylmorpholin-4-yl]quinazolin-2-yl}-1H-pyrazol-1-yl)-2-methylpropan-2-ol, Bromodomain-containing protein 4, GLYCEROL | | 著者 | Nadupalli, A, Fontano, E, Connors, C.R, Chan, S.G, Olland, A.M, Lakshminarasimhan, D, White, A, Suto, R.K. | | 登録日 | 2018-08-28 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Lead optimization and efficacy evaluation of quinazoline-based BET family inhibitors for potential treatment of cancer and inflammatory diseases.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
5IZ7
 
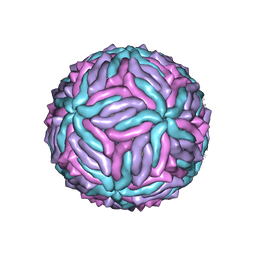 | | Cryo-EM structure of thermally stable Zika virus strain H/PF/2013 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, structural protein E, structural protein M | | 著者 | Kostyuchenko, V.A, Zhang, S, Fibriansah, G, Lok, S.M. | | 登録日 | 2016-03-25 | | 公開日 | 2016-05-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of the thermally stable Zika virus
Nature, 533, 2016
|
|
6IRG
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class II | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (5.5 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRF
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class I | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
