3D8V
 
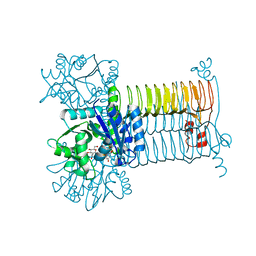 | |
3T7U
 
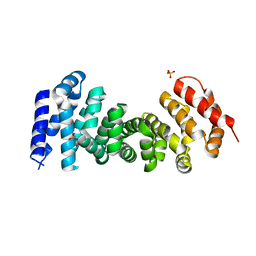 | | A NeW Crystal structure of APC-ARM | | Descriptor: | Adenomatous polyposis coli protein, PHOSPHATE ION | | Authors: | Zhang, Z, Wu, G. | | Deposit date: | 2011-07-31 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the armadillo repeat domain of adenomatous polyposis coli which reveals its inherent flexibility
Biochem.Biophys.Res.Commun., 412, 2011
|
|
3G0C
 
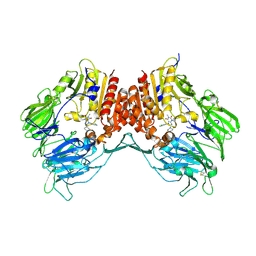 | | Crystal structure of dipeptidyl peptidase IV in complex with a pyrimidinedione inhibitor 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(2-chlorobenzyl)-1,3-dimethyl-8-piperazin-1-yl-3,7-dihydro-1H-purine-2,6-dione, ... | | Authors: | Zhang, Z, Wallace, M.B, Feng, J, Stafford, J.A, Kaldor, S.W, Shi, L, Skene, R.J, Aertgeerts, K, Lee, B, Jennings, A, Xu, R, Kassel, D, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design and Synthesis of Pyrimidinone and Pyrimidinedione Inhibitors of Dipeptidyl Peptidase IV.
J.Med.Chem., 54, 2011
|
|
3G0D
 
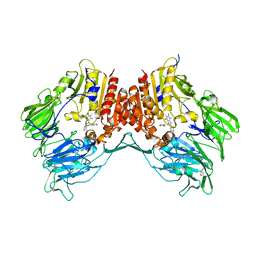 | | Crystal structure of dipeptidyl peptidase IV in complex with a pyrimidinedione inhibitor 2 | | Descriptor: | 2-({8-[(3R)-3-AMINOPIPERIDIN-1-YL]-1,3-DIMETHYL-2,6-DIOXO-1,2,3,6-TETRAHYDRO-7H-PURIN-7-YL}METHYL)BENZONITRILE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Wallace, M.B, Feng, J, Stafford, J.A, Kaldor, S.W, Shi, L, Skene, R.J, Aertgeerts, K, Lee, B, Jennings, A, Xu, R, Kassel, D, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Design and Synthesis of Pyrimidinone and Pyrimidinedione Inhibitors of Dipeptidyl Peptidase IV.
J.Med.Chem., 54, 2011
|
|
3G0B
 
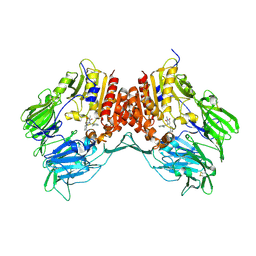 | | Crystal structure of dipeptidyl peptidase IV in complex with TAK-322 | | Descriptor: | 2-({6-[(3R)-3-aminopiperidin-1-yl]-3-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl}methyl)benzonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Wallace, M.B, Feng, J, Stafford, J.A, Kaldor, S.W, Shi, L, Skene, R.J, Aertgeerts, K, Lee, B, Jennings, A, Xu, R, Kassel, D, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design and Synthesis of Pyrimidinone and Pyrimidinedione Inhibitors of Dipeptidyl Peptidase IV.
J.Med.Chem., 54, 2011
|
|
6LVY
 
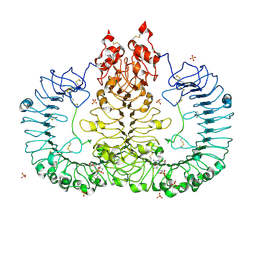 | | Crystal structure of TLR7/Cpd-2 (SM-360320) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-2-(2-methoxyethoxy)-9-(phenylmethyl)-7H-purin-8-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
5Y28
 
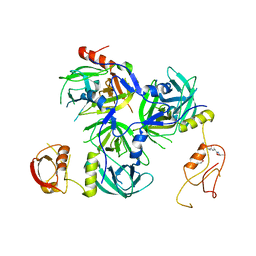 | | Crystal structure of H. pylori HtrA with PDZ2 deletion | | Descriptor: | 1,2-ETHANEDIOL, Periplasmic serine endoprotease DegP-like, UNK-UNK-UNK-UNK | | Authors: | Zhang, Z, Huang, Q, Tao, X. | | Deposit date: | 2017-07-24 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.08606815 Å) | | Cite: | The unique trimeric assembly of the virulence factor HtrA fromHelicobacter pylorioccurs via N-terminal domain swapping.
J.Biol.Chem., 294, 2019
|
|
5Y2D
 
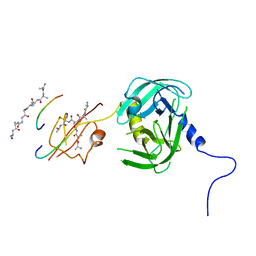 | | Crystal structure of H. pylori HtrA | | Descriptor: | Periplasmic serine endoprotease DegP-like, UNK-UNK-K-UNK-UNK-UNK-UNK-UNK-UNK-UNK, UNK-UNK-UNK, ... | | Authors: | Zhang, Z, Huang, Q, Tao, X. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.70009851 Å) | | Cite: | The unique trimeric assembly of the virulence factor HtrA fromHelicobacter pylorioccurs via N-terminal domain swapping.
J.Biol.Chem., 294, 2019
|
|
5CFF
 
 | | Crystal structure of Miranda/Staufen dsRBD5 complex | | Descriptor: | Miranda, Staufen | | Authors: | Shan, Z, Wen, W. | | Deposit date: | 2015-07-08 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis of Miranda-mediated Staufen localization during Drosophila neuroblast asymmetric division
Nat Commun, 6, 2015
|
|
3JBQ
 
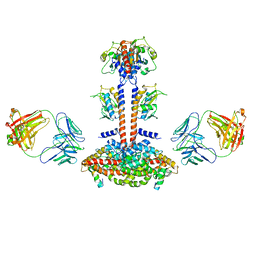 | | Domain Organization and Conformational Plasticity of the G Protein Effector, PDE6 | | Descriptor: | GafA domain of cone phosphodiesterase 6C, GafB domain of phosphodiesterase 2A, IgG1-kappa 2E8 heavy chain, ... | | Authors: | Zhang, Z, He, F, Constantine, R, Baker, M.L, Baehr, W, Schmid, M.F, Wensel, T.G, Agosto, M.A. | | Deposit date: | 2015-09-17 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Domain Organization and Conformational Plasticity of the G Protein Effector, PDE6.
J.Biol.Chem., 290, 2015
|
|
5W81
 
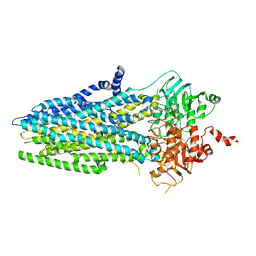 | | Phosphorylated, ATP-bound structure of zebrafish cystic fibrosis transmembrane conductance regulator (CFTR) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Zhang, Z, Liu, F, Chen, J. | | Deposit date: | 2017-06-21 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Conformational Changes of CFTR upon Phosphorylation and ATP Binding.
Cell, 170, 2017
|
|
7U32
 
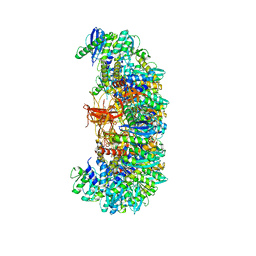 | | MVV cleaved synaptic complex (CSC) intasome at 3.4 A resolution | | Descriptor: | CALCIUM ION, DNA EV272, DNA EV273, ... | | Authors: | Shan, Z, Pye, V.E, Cherepanov, P, Lyumkis, D. | | Deposit date: | 2022-02-25 | | Release date: | 2022-05-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Multivalent interactions essential for lentiviral integrase function.
Nat Commun, 13, 2022
|
|
7E7X
 
 | |
8I96
 
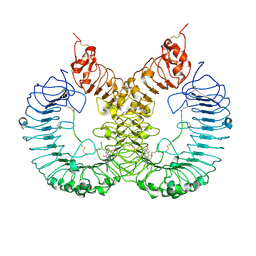 | | Cryo-EM structure of TLR7/TH-407b complex | | Descriptor: | (1S,3R)-5-[4-(8-nitroquinolin-5-yl)piperazin-1-yl]carbonyladamantan-2-one, Toll-like receptor 7 | | Authors: | Zhang, Z. | | Deposit date: | 2023-02-06 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of TLR7/TH-407b complex
To Be Published
|
|
2JX0
 
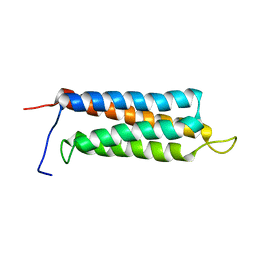 | | The paxillin-binding domain (PBD) of G Protein Coupled Receptor (GPCR)-kinase (GRK) interacting protein 1 (GIT1) | | Descriptor: | ARF GTPase-activating protein GIT1 | | Authors: | Zhang, Z, Guibao, C.D, Simmerman, J.A, Zheng, J. | | Deposit date: | 2007-11-01 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | GIT1 paxillin-binding domain is a four-helix bundle, and it binds to both paxillin LD2 and LD4 motifs.
J.Biol.Chem., 283, 2008
|
|
3JAB
 
 | | Domain organization and conformational plasticity of the G protein effector, PDE6 | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, GafA domain of cone phosphodiesterase 6C, GafB domain of phosphodiesterase 2A, ... | | Authors: | Zhang, Z, He, F, Constantine, R, Baker, M.L, Baehr, W, Schmid, M.F, Wensel, T.G, Agosto, M.A. | | Deposit date: | 2015-05-26 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Domain Organization and Conformational Plasticity of the G Protein Effector, PDE6.
J.Biol.Chem., 290, 2015
|
|
8ZRR
 
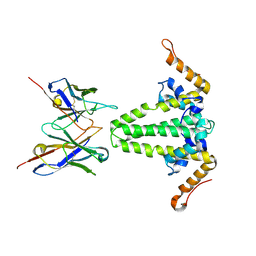 | |
8ZRH
 
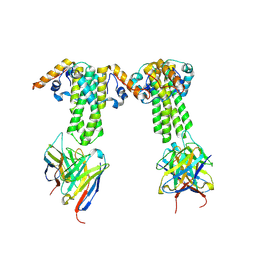 | | HBcAg-D4 Fab complex | | Descriptor: | Capsid protein, Heavy chains of D4 Fab, Light chains of D4 Fab | | Authors: | Zhang, Z, Ju, B, Liu, C, Yan, H. | | Deposit date: | 2024-06-04 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | HBcAg-D4 Fab complex
To Be Published
|
|
2QKX
 
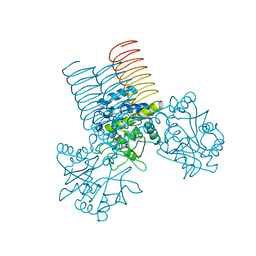 | | N-acetyl glucosamine 1-phosphate uridyltransferase from Mycobacterium tuberculosis complex with N-acetyl glucosamine 1-phosphate | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, Bifunctional protein glmU | | Authors: | Zhang, Z, Squire, C.J, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-07-11 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of GlmU from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2BCC
 
 | | STIGMATELLIN-BOUND CYTOCHROME BC1 COMPLEX FROM CHICKEN | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.I, Kim, K.K, Hung, L.W, Crofts, A.R, Berry, E.A, Kim, S.H. | | Deposit date: | 1998-09-18 | | Release date: | 1999-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
7BZH
 
 | |
7CM5
 
 | | Full-length Sarm1 in a self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
7CM7
 
 | | NAD+-bound Sarm1 E642A in the self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
7CM6
 
 | | NAD+-bound Sarm1 in the self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
2NCA
 
 | |
