2I0I
 
 | | X-ray crystal structure of Sap97 PDZ3 bound to the C-terminal peptide of HPV18 E6 | | Descriptor: | Disks large homolog 1, peptide E6 | | Authors: | Chen, X.S, Zhang, Y, Dasgupta, J, Banks, L, Thomas, M. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of a Human Papillomavirus (HPV) E6 Polypeptide Bound to MAGUK Proteins: Mechanisms of Targeting Tumor Suppressors by a High-Risk HPV Oncoprotein.
J.Virol., 81, 2007
|
|
5X5F
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 2 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
2ITK
 
 | | human Pin1 bound to D-PEPTIDE | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, D-Peptide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Noel, J.P, Zhang, Y. | | Deposit date: | 2006-10-19 | | Release date: | 2007-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for high-affinity peptide inhibition of human Pin1.
Acs Chem.Biol., 2, 2007
|
|
8WM0
 
 | | Crystal structure of TNIK-thiopeptide wTP3 complex | | Descriptor: | ADENOSINE, THIOPEPTIDE wTP3, TRAF2 and NCK-interacting protein kinase | | Authors: | Hamada, K, Kobayashi, S, Vinogradov, A.A, Zhang, Y, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2023-10-01 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Compact Reprogrammed Genetic Code for De Novo Discovery of Proteolytically Stable Thiopeptides.
J.Am.Chem.Soc., 2024
|
|
1FPG
 
 | | STRUCTURAL ASPECTS OF THE ALLOSTERIC INHIBITION OF FRUCTOSE-1,6-BISPHOSPHATASE BY AMP: THE BINDING OF BOTH THE SUBSTRATE ANALOGUE 2,5-ANHYDRO-D-GLUCITOL-1,6-BISPHOSPHATE AND CATALYTIC METAL IONS MONITORED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Villeret, V, Huang, S, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1994-12-15 | | Release date: | 1995-02-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural aspects of the allosteric inhibition of fructose-1,6-bisphosphatase by AMP: the binding of both the substrate analogue 2,5-anhydro-D-glucitol 1,6-bisphosphate and catalytic metal ions monitored by X-ray crystallography.
Biochemistry, 34, 1995
|
|
8P7K
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | (2~{R})-2-methylpentanedioic acid, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Dym, O, Aggawal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-30 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8WY0
 
 | | T cell receptor delta 2 gamma 9 with F283A, F290A, and F291A | | Descriptor: | CHOLESTEROL, Signal peptide,flag tag,T cell receptor delta variable 2,T cell receptor delta constant, Signal peptide,flag tag,T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Liu, Y, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8WYI
 
 | | T cell receptor delta 2 gamma 9 with TCRD TM domain chimera of TRAC | | Descriptor: | Signal peptide,flag tag,T cell receptor delta variable 2,T cell receptor delta constant,T cell receptor alpha chain constant,T cell receptor delta variable 2,T cell receptor delta constant,T cell receptor alpha chain constant, Signal peptide,flag tag,T cell receptor gamma variable 9,T cell receptor gamma constant 1,T cell receptor gamma variable 9,T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Xin, W, Huang, B, Chi, X, Liu, Y, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8WXE
 
 | | Vgamma5Vdelta1 EH TCR-CD3 complex | | Descriptor: | CHOLESTEROL, Signal peptide,flag tag,T cell receptor delta variable 1,T cell receptor delta constant, Signal peptide,flag tag,T cell receptor gamma variable 5,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-10-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
1FPE
 
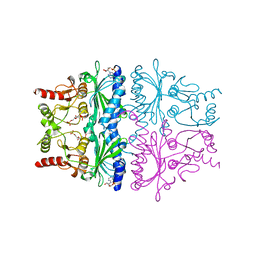 | | STRUCTURAL ASPECTS OF THE ALLOSTERIC INHIBITION OF FRUCTOSE-1,6-BISPHOSPHATASE BY AMP: THE BINDING OF BOTH THE SUBSTRATE ANALOGUE 2,5-ANHYDRO-D-GLUCITOL-1,6-BISPHOSPHATE AND CATALYTIC METAL IONS MONITORED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Villeret, V, Huang, S, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1994-12-15 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural aspects of the allosteric inhibition of fructose-1,6-bisphosphatase by AMP: the binding of both the substrate analogue 2,5-anhydro-D-glucitol 1,6-bisphosphate and catalytic metal ions monitored by X-ray crystallography.
Biochemistry, 34, 1995
|
|
5X4S
 
 | | Structure of the N-terminal domain (NTD)of SARS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
8IVG
 
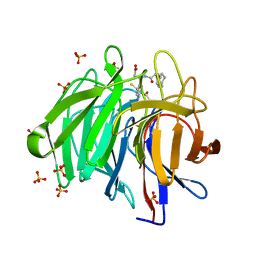 | | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1/Nrf2 PPI Inhibitor for Treatment of Chronic Kidney Disease | | Descriptor: | (3S)-3-(4-methylphenyl)-3-[2-(5,6,7,8-tetrahydronaphthalen-2-yl)ethanoylamino]propanoic acid, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Kelch-like ECH-associated protein 1, ... | | Authors: | Otake, K, Ubukata, M, Nagahashi, N, Ogawa, N, Hantani, Y, Hantani, R, Adachi, T, Nomura, A, Yamaguchi, K, Maekawa, M, Mamada, H, Motomura, T, Sato, M, Harada, K. | | Deposit date: | 2023-03-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1-Nrf2 PPI Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
1ZCN
 
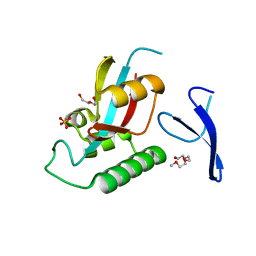 | | human Pin1 Ng mutant | | Descriptor: | PENTAETHYLENE GLYCOL, PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jager, M, Zhang, Y, Nguyen, H, Dendel, G, Bowman, M.E, Gruebele, M, Noel, J.P, Kelly, J.W. | | Deposit date: | 2005-04-12 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function-folding relationship in a WW domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
8JFQ
 
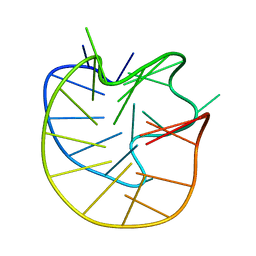 | | Structure of the Major G-Quadruplex in the Human EGFR Oncogene Promoter Adopts a Unique Folding Topology with a Distinctive Snap-back Loop | | Descriptor: | 26mer-DNA | | Authors: | Liu, Y, Li, J, Zhang, Y, Wang, Y, Chen, J, Bian, Y, Xia, Y, Yang, M.H, Zheng, K, Wang, K.B, Kong, L.Y. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Major G-Quadruplex in the Human EGFR Oncogene Promoter Adopts a Unique Folding Topology with a Distinctive Snap-Back Loop.
J.Am.Chem.Soc., 145, 2023
|
|
3JSF
 
 | |
5FBP
 
 | | CRYSTAL STRUCTURE OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH THE PRODUCT FRUCTOSE 6-PHOSPHATE AT 2.1-ANGSTROMS RESOLUTION | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Ke, H, Liang, J.-Y, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1991-02-11 | | Release date: | 1992-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the neutral form of fructose-1,6-bisphosphatase complexed with the product fructose 6-phosphate at 2.1-A resolution.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
2CEU
 
 | | Despentapeptide insulin in acetic acid (pH 2) | | Descriptor: | INSULIN, SULFATE ION | | Authors: | Whittingham, J.L, Zhang, Y, Zakova, L, Dodson, E.J, Turkenburg, J.P, Brange, J, Dodson, G.G. | | Deposit date: | 2006-02-10 | | Release date: | 2006-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | I222 Crystal Form of Despentapeptide (B26-B30) Insulin Provides New Insights Into the Properties of Monomeric Insulin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5X4R
 
 | | Structure of the N-terminal domain (NTD) of MERS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X58
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5Y93
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[5-[(5-bromanyl-2-methoxy-phenyl)sulfonylamino]-3-methyl-1,2-benzoxazol-6-yl]oxy]-N-(2-morpholin-4-ylethyl)ethanamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
1NYP
 
 | | 4th LIM domain of PINCH protein | | Descriptor: | PINCH protein, ZINC ION | | Authors: | Velyvis, A, Vaynberg, J, Vinogradova, O, Zhang, Y, Wu, C, Qin, J. | | Deposit date: | 2003-02-13 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into PINCH LIM4 domain-mediated integrin signaling
Nat.Struct.Biol., 10, 2003
|
|
4EAC
 
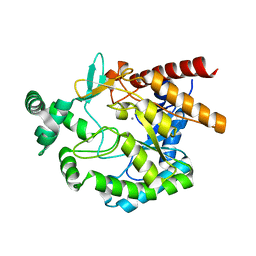 | | Crystal structure of mannonate dehydratase from Escherichia coli strain K12 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Qiu, X, Zhu, Y, Yuan, Y, Zhang, Y, Liu, H, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2012-03-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into decreased enzymatic activity induced by an insert sequence in mannonate dehydratase from Gram negative bacterium.
J.Struct.Biol., 180, 2012
|
|
1Z68
 
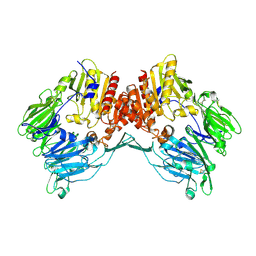 | | Crystal Structure Of Human Fibroblast Activation Protein alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aertgeerts, K, Levin, I, Shi, L, Prasad, G.S, Zhang, Y, Kraus, M.L, Salakian, S, Snell, G.P, Sridhar, V, Wijnands, R, Tennant, M.G. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and kinetic analysis of the substrate specificity of human fibroblast activation protein alpha.
J.Biol.Chem., 280, 2005
|
|
5X5B
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 2 | | Descriptor: | Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
