6VGS
 
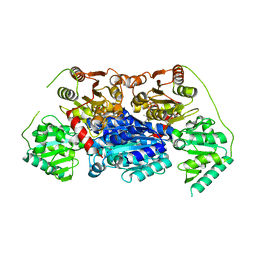 | | Alpha-ketoisovalerate decarboxylase (KivD) from Lactococcus lactis, thermostable mutant | | Descriptor: | Alpha-keto acid decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Chan, S, Korman, T.P, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2020-01-08 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isobutanol production freed from biological limits using synthetic biochemistry.
Nat Commun, 11, 2020
|
|
6YED
 
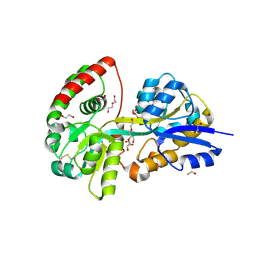 | | E.coli's Putrescine receptor PotF in its open apo state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
3L6P
 
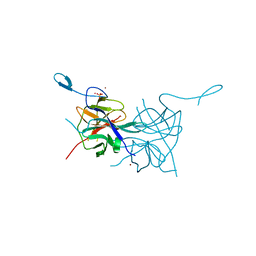 | | Crystal Structure of Dengue Virus 1 NS2B/NS3 protease | | Descriptor: | CADMIUM ION, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Chandramouli, S, Joseph, J.S, Daudenarde, S, Gatchalian, J, Cornillez-Ty, C, Kuhn, P. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Serotype-specific structural differences in the protease-cofactor complexes of the dengue virus family.
J.Virol., 84, 2010
|
|
2DWD
 
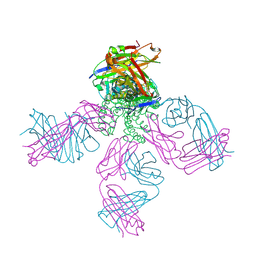 | | crystal structure of KcsA-FAB-TBA complex in Tl+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, ANTIBODY FAB HEAVY CHAIN, ANTIBODY FAB LIGHT CHAIN, ... | | Authors: | Yohannan, S, Zhou, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel
J.Mol.Biol., 366, 2007
|
|
6Y4S
 
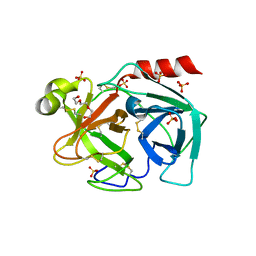 | |
6SJU
 
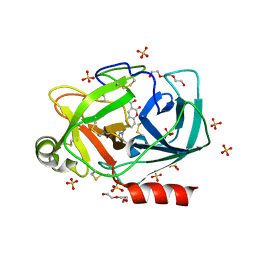 | |
2DWE
 
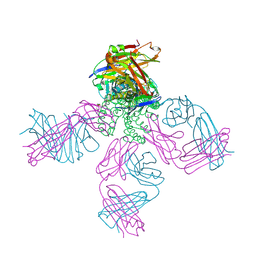 | | Crystal structure of KcsA-FAB-TBA complex in Rb+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, ANTIBODY FAB HEAVY CHAIN, ANTIBODY FAB LIGHT CHAIN, ... | | Authors: | Yohannan, S, Zhou, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel
J.Mol.Biol., 366, 2007
|
|
6SHI
 
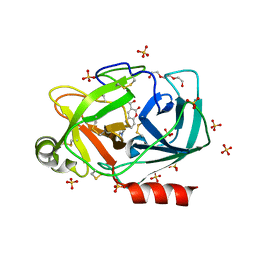 | |
6YE7
 
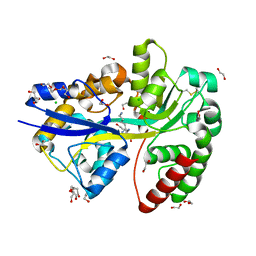 | | E.coli's Putrescine receptor PotF complexed with Cadaverine | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
1S54
 
 | | Thr24Ala Bacteriorhodopsin | | Descriptor: | RETINAL, bacteriorhodopsin | | Authors: | Yohannan, S, Faham, S, Yang, D, Grosfeld, D, Chamberlain, A.K, Bowie, J.U. | | Deposit date: | 2004-01-19 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A C(alpha)-H.O Hydrogen Bond in a Membrane Protein Is Not Stabilizing
J.Am.Chem.Soc., 126, 2004
|
|
1S53
 
 | | Thr46Ser Bacteriorhodopsin | | Descriptor: | RETINAL, bacteriorhodopsin | | Authors: | Yohannan, S, Faham, S, Yang, D, Grosfeld, D, Chamberlain, A.K, Bowie, J.U. | | Deposit date: | 2004-01-19 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A C(alpha)-H.O Hydrogen Bond in a Membrane Protein Is Not Stabilizing
J.Am.Chem.Soc., 126, 2004
|
|
1S52
 
 | | Thr24Val Bacteriorhodopsin | | Descriptor: | RETINAL, bacteriorhodopsin | | Authors: | Yohannan, S, Faham, S, Yang, D, Grosfeld, D, Chamberlain, A.K, Bowie, J.U. | | Deposit date: | 2004-01-19 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A C(alpha)-H.O Hydrogen Bond in a Membrane Protein Is Not Stabilizing
J.Am.Chem.Soc., 126, 2004
|
|
5E59
 
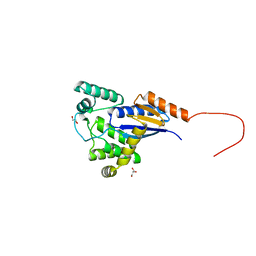 | |
1RA0
 
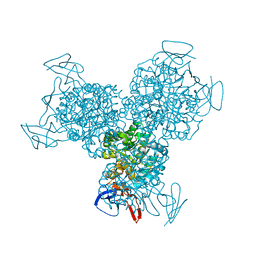 | | Bacterial cytosine deaminase D314G mutant bound to 5-fluoro-4-(S)-hydroxy-3,4-dihydropyrimidine. | | Descriptor: | (4S)-5-FLUORO-4-HYDROXY-3,4-DIHYDROPYRIMIDIN-2(1H)-ONE, Cytosine deaminase, FE (III) ION | | Authors: | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
2GAB
 
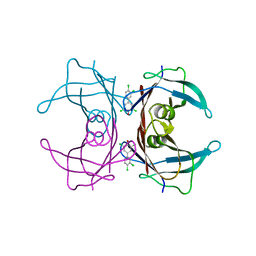 | | Human Transthyretin (TTR) Complexed with Hydroxylated polychlorinated Biphenyl-4-hydroxy-3,3',5,4'-tetrachlorobiphenyl | | Descriptor: | 3,3',4',5-TETRACHLOROBIPHENYL-4-OL, Transthyretin | | Authors: | Palaninathan, S.K, Smith, C, Safe, S.H, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2006-03-08 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Hydroxylated polychlorinated biphenyls selectively bind transthyretin in blood and inhibit amyloidogenesis: rationalizing rodent PCB toxicity
Chem.Biol., 11, 2004
|
|
1RA5
 
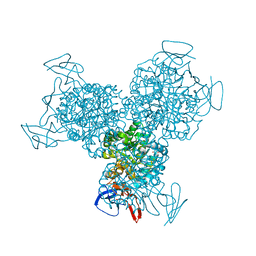 | | Bacterial cytosine deaminase D314A mutant bound to 5-fluoro-4-(S)-hydroxyl-3,4-dihydropyrimidine. | | Descriptor: | (4S)-5-FLUORO-4-HYDROXY-3,4-DIHYDROPYRIMIDIN-2(1H)-ONE, Cytosine deaminase, FE (III) ION, ... | | Authors: | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
6H45
 
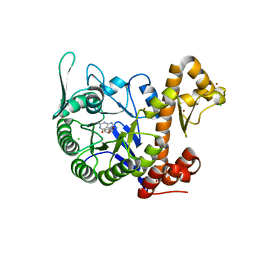 | | crystal structure of the human TGT catalytic subunit QTRT1 in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Johannsson, S, Neumann, P, Ficner, R. | | Deposit date: | 2018-07-20 | | Release date: | 2018-09-05 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human tRNA Guanine Transglycosylase Catalytic Subunit QTRT1.
Biomolecules, 8, 2018
|
|
6SHH
 
 | |
1RAK
 
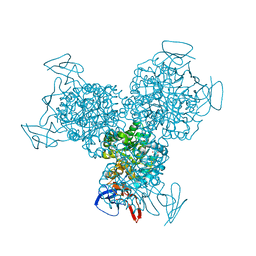 | | Bacterial cytosine deaminase D314S mutant bound to 5-fluoro-4-(S)-hydroxyl-3,4-dihydropyrimidine. | | Descriptor: | (4S)-5-FLUORO-4-HYDROXY-3,4-DIHYDROPYRIMIDIN-2(1H)-ONE, Cytosine deaminase, FE (III) ION, ... | | Authors: | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
5AEI
 
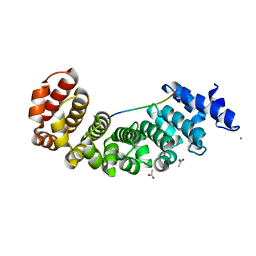 | | Designed Armadillo repeat protein YIIIM5AII in complex with peptide (KR)5 | | Descriptor: | ACETATE ION, CALCIUM ION, DESIGNED ARMADILLO REPEAT PROTEIN YIIIM5AII, ... | | Authors: | Hansen, S, Tremmel, D, Madhurantakam, C, Reichen, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2015-08-31 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and Energetic Contributions of a Designed Modular Peptide-Binding Protein with Picomolar Affinity.
J.Am.Chem.Soc., 138, 2016
|
|
2BYN
 
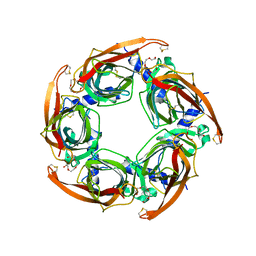 | | Crystal structure of apo AChBP from Aplysia californica | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PENTAETHYLENE GLYCOL, SOLUBLE ACETYLCHOLINE RECEPTOR, ... | | Authors: | Hansen, S.B, Sulzenbacher, G, Huxford, T, Marchot, P, Taylor, P, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structures of Aplysia Achbp Complexes with Nicotinic Agonists and Antagonists Reveal Distinctive Binding Interfaces and Conformations.
Embo J., 24, 2005
|
|
5BN5
 
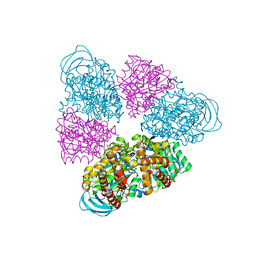 | | Structural basis for a unique ATP synthase core complex from Nanoarcheaum equitans | | Descriptor: | NEQ263, SULFATE ION, V-type ATP synthase alpha chain | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
2JRK
 
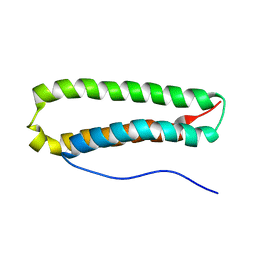 | | NMR Structure and Epitope Mapping of Blo t 5 | | Descriptor: | Mite allergen Blo t 5 | | Authors: | Chan, S, Mok, Y. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure and IgE epitopes of Blo t 5, a major dust mite allergen
J.Immunol., 181, 2008
|
|
1TN5
 
 | | Structure of bacterorhodopsin mutant K41P | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Yohannan, S, Yang, D, Faham, S, Boulting, G, Whitelegge, J, Bowie, J.U. | | Deposit date: | 2004-06-11 | | Release date: | 2004-10-19 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Proline substitutions are not easily accommodated in a membrane protein
J.Mol.Biol., 341, 2004
|
|
5BN3
 
 | | Structure of a unique ATP synthase NeqA-NeqB in complex with ADP from Nanoarcheaum equitans | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
