4P1V
 
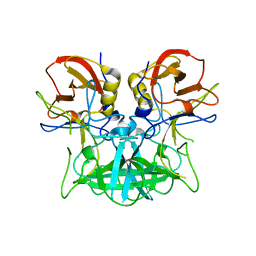 | | Structure of the P domain from a GI.7 Norovirus variant in complex with H-type 2 HBGA | | Descriptor: | P domain of VPI, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-02-27 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5497 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
1HNB
 
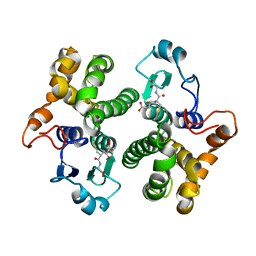 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
1HNC
 
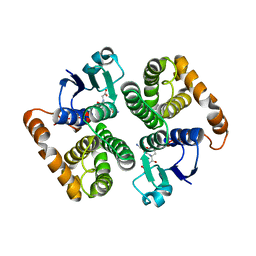 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
1JPJ
 
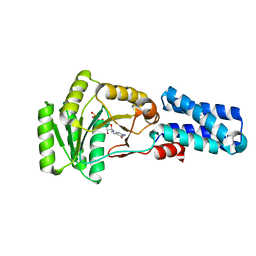 | | GMPPNP Complex of SRP GTPase NG Domain | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, SIGNAL RECOGNITION PARTICLE PROTEIN | | Authors: | Padmanabhan, S, Freymann, D.M. | | Deposit date: | 2001-08-02 | | Release date: | 2002-02-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The conformation of bound GMPPNP suggests a mechanism for gating the active site of the SRP GTPase.
Structure, 9, 2001
|
|
1JV6
 
 | | BACTERIORHODOPSIN D85S/F219L DOUBLE MUTANT AT 2.00 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Rouhani, S, Cartailler, J.-P, Facciotti, M.T, Walian, P, Needleman, R, Lanyi, J.K, Glaeser, R.M, Luecke, H. | | Deposit date: | 2001-08-28 | | Release date: | 2001-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the D85S mutant of bacteriorhodopsin: model of an O-like photocycle intermediate.
J.Mol.Biol., 313, 2001
|
|
1BXN
 
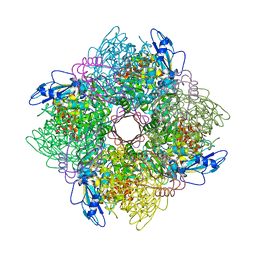 | | THE CRYSTAL STRUCTURE OF RUBISCO FROM ALCALIGENES EUTROPHUS TO 2.7 ANGSTROMS. | | Descriptor: | PHOSPHATE ION, PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN), PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL CHAIN) | | Authors: | Hansen, S, Vollan, V.B, Hough, E, Andersen, K. | | Deposit date: | 1998-10-06 | | Release date: | 1999-10-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of rubisco from Alcaligenes eutrophus reveals a novel central eight-stranded beta-barrel formed by beta-strands from four subunits.
J.Mol.Biol., 288, 1999
|
|
8BYR
 
 | |
4L2W
 
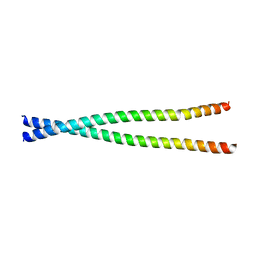 | |
3Q4H
 
 | | Crystal structure of the Mycobacterium smegmatis EsxGH complex (MSMEG_0620-MSMEG_0621) | | Descriptor: | Low molecular weight protein antigen 7, Pe family protein | | Authors: | Chan, S, Harris, L, Kuo, E, Ahn, C, Zhou, T.T, Nguyen, L, Shin, A, Sawaya, M.R, Cascio, D, Arbing, M.A, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-12-23 | | Release date: | 2011-01-26 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
8WMH
 
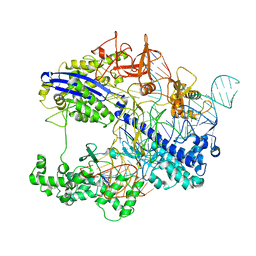 | | Structure of CbCas9 bound to 6-nucleotide complementary DNA substrate | | Descriptor: | NTS, TS, deadCbCas9, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-03 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WMM
 
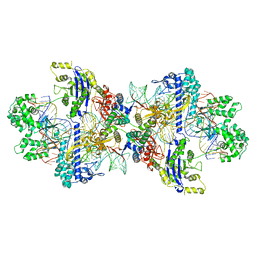 | | Structure of CbCas9-PcrIIC1 complex bound to 28-bp DNA substrate (20-nt complementary) | | Descriptor: | MAGNESIUM ION, NTS, PcrIIC1, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WMN
 
 | | Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (symmetric 20-nt complementary) | | Descriptor: | DNA (62-MER), MAGNESIUM ION, PcrIIC1, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WR4
 
 | | Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (non-targeting complex) | | Descriptor: | CbCas9 effector-1, DNA (62-MER), MAGNESIUM ION, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-13 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
3K6V
 
 | | M. acetivorans Molybdate-Binding Protein (ModA) in Citrate-Bound Open Form | | Descriptor: | CITRIC ACID, Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4P3I
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeA HBGA. | | Descriptor: | P domain of VP1, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6941 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P12
 
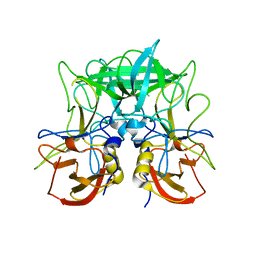 | | Native Structure of the P domain from a GI.7 Norovirus variant. | | Descriptor: | Major capsid protein | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-02-24 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6011 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4JFA
 
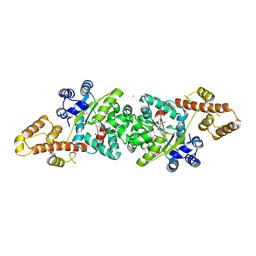 | | Crystal Structure of Plasmodium falciparum Tryptophanyl-tRNA synthetase | | Descriptor: | BETA-MERCAPTOETHANOL, POTASSIUM ION, TRYPTOPHAN, ... | | Authors: | Khan, S, Garg, A, Manickam, Y, Sharma, A. | | Deposit date: | 2013-02-28 | | Release date: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An appended domain results in an unusual architecture for malaria parasite tryptophanyl-tRNA synthetase
Plos One, 8, 2013
|
|
4P25
 
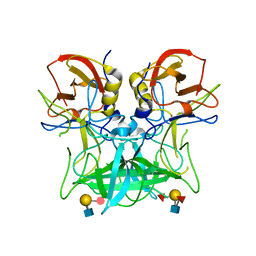 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeY HBGA. | | Descriptor: | Major capsid protein, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4991 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P2N
 
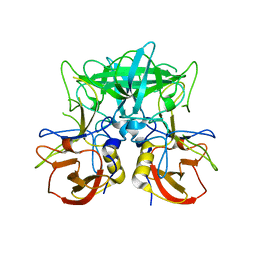 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeX HBGA | | Descriptor: | Major capsid protein, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
3JRN
 
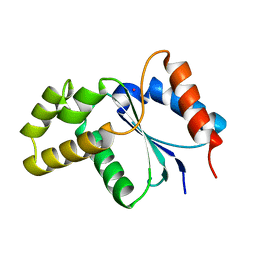 | | Crystal structure of TIR domain from Arabidopsis Thaliana | | Descriptor: | ARSENIC, AT1G72930 protein | | Authors: | Chan, S.L, Mukasa, T, Santelli, E, Low, L.Y, Pascual, J. | | Deposit date: | 2009-09-08 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a TIR domain from Arabidopsis thaliana reveals a conserved helical region unique to plants.
Protein Sci., 19, 2009
|
|
4PDQ
 
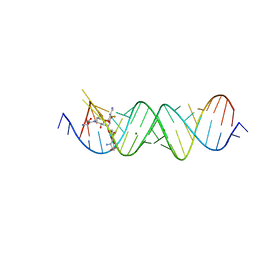 | | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neomycin analog | | Descriptor: | (2S)-4-amino-N-{(1R,2S,3R,4R,5S)-5-amino-3-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy }-4-[(2,6-diamino-2,4,6-trideoxy-4-fluoro-alpha-D-galactopyranosyl)oxy]-2-hydroxycyclohexyl}-2-hydroxybutanamide, MAGNESIUM ION, RNA (5'-*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C-3') | | Authors: | Hanessian, S, Saavedra, O.M, Vilchis-Reyes, M.A, Maianti, J.P, Kanazawa, H, Dozzo, P, Feeney, L.A, Armstrong, E.S, Kondo, J. | | Deposit date: | 2014-04-21 | | Release date: | 2015-01-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis, broad spectrum antibacterial activity, and X-ray co-crystal structure of the decoding bacterial ribosomal A-site with 4'-deoxy-4'-fluoro neomycin analogs
Chem Sci, 5, 2014
|
|
2L5X
 
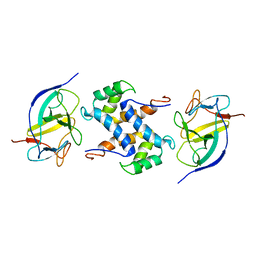 | | Solution structure of IL1A-S100A13 complex | | Descriptor: | Interleukin-1 alpha, Protein S100-A13 | | Authors: | Mohan, S.K, Yu, C. | | Deposit date: | 2010-11-09 | | Release date: | 2011-01-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The IL1alpha-S100A13 heterotetrameric complex structure: a component in the non-classical pathway for interleukin 1alpha secretion
J.Biol.Chem., 286, 2011
|
|
1EYG
 
 | |
2K43
 
 | |
3K6X
 
 | | M. acetivorans Molybdate-Binding Protein (ModA) in Molybdate-Bound Close Form with 2 Molecules in Asymmetric Unit Forming Beta Barrel | | Descriptor: | MOLYBDATE ION, SULFATE ION, Solute-binding protein MA_0280 | | Authors: | Chan, S, Chernishof, I, Giuroiu, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
