6QJE
 
 | | Structure of human galactokinase 1 bound with 4-{[2-(Methylsulfonyl)-1H-imidazol-1-yl]methyl}-1,3-thiazole | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, 4-[(2-methylsulfonylimidazol-1-yl)methyl]-1,3-thiazole, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with 4-{[2-(Methylsulfonyl)-1H-imidazol-1-yl]methyl}-1,3-thiazole
To Be Published
|
|
6Q90
 
 | | Structure of human galactokinase 1 bound with 1-(4-Methoxyphenyl)-3-(4-pyridinyl)urea | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, N-(4-methoxyphenyl)-N'-pyridin-4-ylurea, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-16 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with 1-(4-Methoxyphenyl)-3-(4-pyridinyl)urea
To Be Published
|
|
3ZJT
 
 | | Ternary complex of E.coli leucyl-tRNA synthetase, tRNA(Leu)574 and the benzoxaborole AN3017 in the editing conformation | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZJV
 
 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3213 in the editing conformation | | Descriptor: | LEUCINE--TRNA LIGASE, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZJU
 
 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3016 in the editing conformation | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
7YUY
 
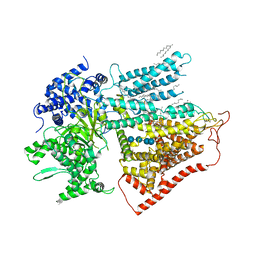 | | Structure of a mutated membrane-bound glycosyltransferase | | Descriptor: | (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, X.L, Yang, P, Zhang, M, Liu, X.T, Yu, H.J. | | Deposit date: | 2022-08-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and mechanistic insights into fungal beta-1,3-glucan synthase FKS1.
Nature, 616, 2023
|
|
6LYW
 
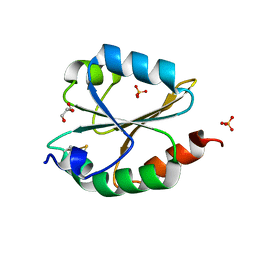 | | Structural insight into the biological functions of Arabidopsis thaliana ACHT1 | | Descriptor: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | Authors: | Wang, J.C, Pan, W.M, Wang, M.Z, Zhang, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
6LYX
 
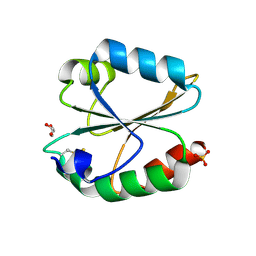 | | Crystal structure of oxidized ACHT1 | | Descriptor: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | Authors: | Wang, J.C, Pan, W.M, Cai, W.G, Wang, M.Z, Zhang, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
5ZRX
 
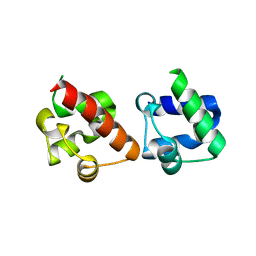 | | Crystal Structure of EphA2/SHIP2 Complex | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2,Ephrin type-A receptor 2 | | Authors: | Wang, Y, Shang, Y, Li, J, Chen, W, Li, G, Wan, J, Liu, W, Zhang, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.
Elife, 7, 2018
|
|
5ZRY
 
 | | Crystal Structure of EphA6/Odin Complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Ankyrin repeat and SAM domain-containing protein 1A,Ephrin type-A receptor 6, ... | | Authors: | Wang, Y, Shang, Y, Li, J, Chen, W, Li, G, Wan, J, Liu, W, Zhang, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.
Elife, 7, 2018
|
|
4DC2
 
 | | Structure of PKC in Complex with a Substrate Peptide from Par-3 | | Descriptor: | ADENINE, Partitioning defective 3 homolog, Protein kinase C iota type | | Authors: | Shang, Y, Wang, C, Yu, J, Zhang, M. | | Deposit date: | 2012-01-17 | | Release date: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate recognition mechanism of atypical protein kinase Cs revealed by the structure of PKC iota in complex with a substrate peptide from Par-3
Structure, 20, 2012
|
|
5ZRZ
 
 | | Crystal Structure of EphA5/SAMD5 Complex | | Descriptor: | Ephrin type-A receptor 5, Sterile alpha motif domain-containing protein 5 | | Authors: | Wang, Y, Shang, Y, Li, J, Chen, W, Li, G, Wan, J, Liu, W, Zhang, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.
Elife, 7, 2018
|
|
7BHS
 
 | | Crystal structure of MAT2a with quinazoline fragment 2 bound in the allosteric site | | Descriptor: | 6-chloranyl-2-methoxy-4-phenyl-quinazoline, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHU
 
 | | Crystal structure of MAT2a with elaborated fragment 26 bound in the allosteric site | | Descriptor: | 1,2-ETHANEDIOL, 7-chloranyl-4-(dimethylamino)-1-(2-hydroxyethyl)quinazolin-2-one, S-ADENOSYLMETHIONINE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHW
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 29) | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-(3-methylphenyl)quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHR
 
 | | Crystal structure of MAT2a with triazinone fragment 1 bound in the allosteric site | | Descriptor: | 4-(dimethylamino)-6-ethoxy-1~{H}-1,3,5-triazin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHV
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor and in vivo tool compound 28 | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-phenyl-quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHT
 
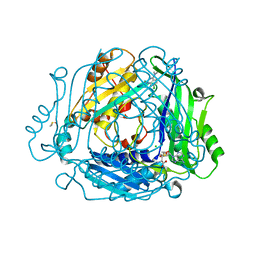 | | Crystal structure of MAT2a with quinazolinone fragment 5 bound in the allosteric site | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1~{H}-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.052 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHX
 
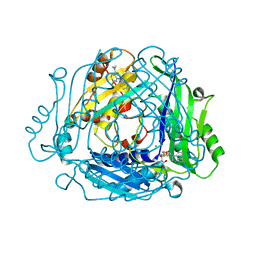 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 31) | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-pyridin-3-yl-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
4EHQ
 
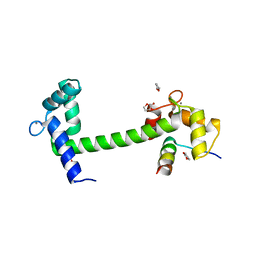 | | Crystal Structure of Calmodulin Binding Domain of Orai1 in Complex with Ca2+/Calmodulin Displays a Unique Binding Mode | | Descriptor: | CALCIUM ION, Calcium release-activated calcium channel protein 1, Calmodulin, ... | | Authors: | Liu, Y, Zheng, X, Mueller, G.A, Sobhany, M, DeRose, E.F, Zhang, Y, London, R.E, Birnbaumer, L. | | Deposit date: | 2012-04-03 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9005 Å) | | Cite: | Crystal structure of calmodulin binding domain of orai1 in complex with ca2+*calmodulin displays a unique binding mode.
J.Biol.Chem., 287, 2012
|
|
7VI7
 
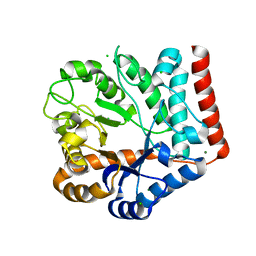 | | Crystal structure of GH3 beta-N-acetylhexosaminidase Amuc_2109 from Akkermansia muciniphila in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Qian, K, Yang, W, Chen, X, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2021-09-26 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of a GH3 beta-N-acetylhexosaminidase from Akkermansia muciniphila involved in mucin degradation
Biochem.Biophys.Res.Commun., 589, 2022
|
|
7VI6
 
 | | Crystal structure of GH3 beta-N-acetylhexosaminidase Amuc_2109 from Akkermansia muciniphila | | Descriptor: | Beta-N-acetylhexosaminidase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Qian, K, Yang, W, Chen, X, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2021-09-26 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of a GH3 beta-N-acetylhexosaminidase from Akkermansia muciniphila involved in mucin degradation
Biochem.Biophys.Res.Commun., 589, 2022
|
|
7F7G
 
 | | a linear Peptide Inhibitors in complex with GK domain | | Descriptor: | DLG4 GK domain, UNK-ARG-ILE-ARG-ARG-ASP-GLU-TYR-LEU-LYS-ALA-ILE-GLN-UNK | | Authors: | Shang, Y, Huang, X, Li, X, Zhang, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Rsc Chem Biol, 2, 2021
|
|
7F7I
 
 | | Stapled Peptide Inhibitor in complex with PSD95 GK domain | | Descriptor: | ACE-ARG-ILE-ARG-ARG-ASP-GLU-TYR-LEU-LYZ-ALA-ILE-GLN-NH2, Disks large homolog 4 | | Authors: | Shang, Y, Huang, X, Li, X, Zhang, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Rsc Chem Biol, 2, 2021
|
|
3EPU
 
 | | Crystal Structure of STM2138, a novel virulence chaperone in Salmonella | | Descriptor: | STM2138 Virulence Chaperone | | Authors: | Zhang, K, Andres, S.N, Hannemann, M, Coombes, B, Junop, M. | | Deposit date: | 2008-09-30 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis and quantitative proteomic
interactome of a novel virulence chaperone in Salmonella
To be Published
|
|
