5XPD
 
 | |
6FV1
 
 | | Structure of human coronavirus NL63 main protease in complex with the alpha-ketoamide (S)-N-((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)-2-cinnamamido-4-methylpentanamide (cinnamoyl-leucine-GlnLactam-CO-CO-NH-benzyl) | | 分子名称: | (2~{S})-4-methyl-~{N}-[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pentanamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | 著者 | Zhang, L, Hilgenfeld, R. | | 登録日 | 2018-02-28 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Alpha-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication Structure-based design, synthesis, and activity assessment.
J.Med.Chem., 2020
|
|
7N6G
 
 | | C1 of central pair | | 分子名称: | CPC1, Calmodulin, DPY30, ... | | 著者 | Han, L, Zhang, K. | | 登録日 | 2021-06-08 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-12-25 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7N61
 
 | | structure of C2 projections and MIPs | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, FAP147, FAP178, ... | | 著者 | Han, L, Zhang, K. | | 登録日 | 2021-06-07 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1HQ3
 
 | | CRYSTAL STRUCTURE OF THE HISTONE-CORE-OCTAMER IN KCL/PHOSPHATE | | 分子名称: | CHLORIDE ION, HISTONE H2A-IV, HISTONE H2B, ... | | 著者 | Chantalat, L, Nicholson, J.M, Lambert, S.J, Reid, A.J, Donovan, M.J, Reynolds, C.D, Wood, C.M, Baldwin, J.P. | | 登録日 | 2000-12-14 | | 公開日 | 2001-01-24 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure of the histone-core octamer in KCl/phosphate crystals at 2.15 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6UVN
 
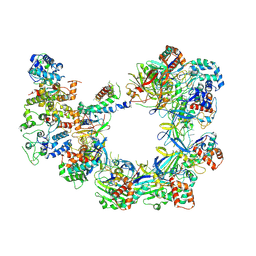 | | CryoEM structure of VcCascasde-TniQ complex | | 分子名称: | Cas6, Cas7, Cas8/5, ... | | 著者 | Chang, L, Li, Z, Zhang, H. | | 登録日 | 2019-11-03 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM structure of a type I-F CRISPR RNA guided surveillance complex bound to transposition protein TniQ.
Cell Res., 30, 2020
|
|
7OQ6
 
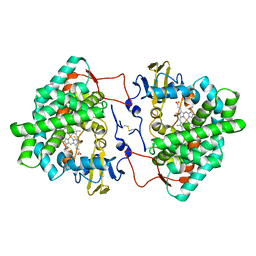 | | Crystal structure of cytochrome P450 Sas16 from Streptomyces asterosporus | | 分子名称: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, THIOCYANATE ION | | 著者 | Zhang, L, Zhang, S, Bechthold, A, Einsle, O. | | 登録日 | 2021-06-02 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | P450-mediated dehydrotyrosine formation during WS9326 biosynthesis proceeds via dehydrogenation of a specific acylated dipeptide substrate.
Acta Pharm Sin B, 13, 2023
|
|
8V32
 
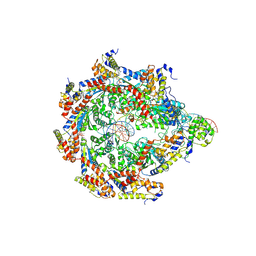 | | TnsD-TnsC-DNA complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA (41-MER), MAGNESIUM ION, ... | | 著者 | Chang, L, Wang, S. | | 登録日 | 2023-11-26 | | 公開日 | 2024-10-16 | | 最終更新日 | 2025-05-14 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Structure of TnsABCD transpososome reveals mechanisms of targeted DNA transposition.
Cell, 187, 2024
|
|
7N3O
 
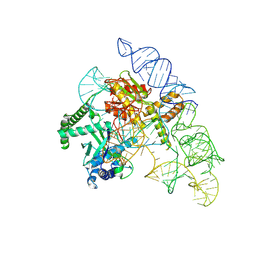 | | Cryo-EM structure of the Cas12k-sgRNA complex | | 分子名称: | Cas12k, Single guide RNA | | 著者 | Chang, L, Li, Z, Xiao, R, Wang, S, Han, R. | | 登録日 | 2021-06-01 | | 公開日 | 2021-09-01 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of target DNA recognition by CRISPR-Cas12k for RNA-guided DNA transposition.
Mol.Cell, 81, 2021
|
|
7N3P
 
 | | Cryo-EM structure of the Cas12k-sgRNA-dsDNA complex | | 分子名称: | Cas12k, DNA (5'-D(*CP*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*AP*AP*CP*CP*GP*AP*GP*TP*TP*T)-3'), DNA (5'-D(P*AP*AP*AP*CP*TP*CP*GP*GP*TP*T)-3'), ... | | 著者 | Chang, L, Li, Z, Xiao, R, Wang, S, Han, R. | | 登録日 | 2021-06-01 | | 公開日 | 2021-09-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Structural basis of target DNA recognition by CRISPR-Cas12k for RNA-guided DNA transposition.
Mol.Cell, 81, 2021
|
|
7WOL
 
 | | Crystal structure of lipase TrLipB from Thermomocrobium roseum | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Zhang, L, Fang, Y, Shi, Y, Gu, Z, Xin, Y. | | 登録日 | 2022-01-21 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of lipase TrLipB from Thermomocrobium roseum
To Be Published
|
|
7N7V
 
 | | Crystal structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes at 2 A. | | 分子名称: | CHLORIDE ION, FE (II) ION, Predicted hydroxylase | | 著者 | Han, L, Xu, W, Ma, M, Miller, M.D, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2021-06-11 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes.
To Be Published
|
|
5U8M
 
 | |
7YSE
 
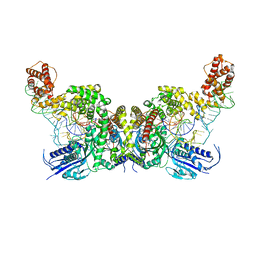 | | Crystal structure of E. coli heterotetrameric GlyRS in complex with tRNA | | 分子名称: | Glycine--tRNA ligase alpha subunit, Glycine--tRNA ligase beta subunit, MAGNESIUM ION, ... | | 著者 | Han, L, Ju, Y, Zhou, H. | | 登録日 | 2022-08-12 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.907 Å) | | 主引用文献 | The binding mode of orphan glycyl-tRNA synthetase with tRNA supports the synthetase classification and reveals large domain movements.
Sci Adv, 9, 2023
|
|
6DA9
 
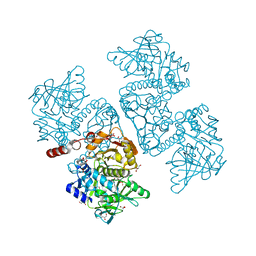 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with FMN bound at 2.05 A resolution | | 分子名称: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Xu, W, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2018-05-01 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
5HOQ
 
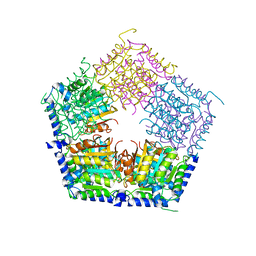 | | Apo structure of CalS11, TDP-rhamnose 3'-o-methyltransferase, an enzyme in Calicheamicin biosynthesis | | 分子名称: | SULFATE ION, TDP-rhamnose 3'-O-methyltransferase (CalS11) | | 著者 | Han, L, Helmich, K.E, Singh, S, Thorson, J.S, Bingman, C.A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis | | 登録日 | 2016-01-19 | | 公開日 | 2016-03-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.793 Å) | | 主引用文献 | Loop dynamics of thymidine diphosphate-rhamnose 3'-O-methyltransferase (CalS11), an enzyme in calicheamicin biosynthesis.
Struct Dyn., 3, 2016
|
|
5VFA
 
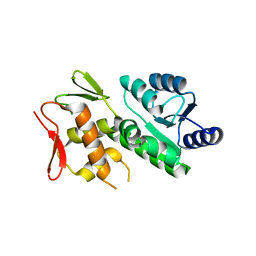 | | RitR Mutant - C128D | | 分子名称: | Response regulator | | 著者 | Han, L, Silvaggi, N.R. | | 登録日 | 2017-04-07 | | 公開日 | 2018-04-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.452 Å) | | 主引用文献 | RitR is an archetype for a novel family of redox sensors in the streptococci that has evolved from two-component response regulators and is required for pneumococcal colonization.
PLoS Pathog., 14, 2018
|
|
6DA6
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes, apo form at 2.6 A resolution (P212121) | | 分子名称: | GLYCEROL, MAGNESIUM ION, UNKNOWN LIGAND, ... | | 著者 | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2018-05-01 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6DA7
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with apo form at 1.83 A resolution (I222) | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | 著者 | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2018-05-01 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6C8T
 
 | | The structure of MppP soaked with the substrate L-Arg | | 分子名称: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, CHLORIDE ION, PLP-Dependent L-Arginine Hydroxylase MppP | | 著者 | Han, L, Silvaggi, N.R. | | 登録日 | 2018-01-25 | | 公開日 | 2018-09-05 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
5BK7
 
 | |
6C92
 
 | | The structure of MppP soaked with the product 2-ketoarginine | | 分子名称: | (4S)-5-carbamimidamido-4-hydroxy-2-oxopentanoic acid, (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, PLP-Dependent L-Arginine Hydroxylase MppP | | 著者 | Han, L, Silvaggi, N.R. | | 登録日 | 2018-01-25 | | 公開日 | 2018-09-05 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.834 Å) | | 主引用文献 | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
6C9B
 
 | |
4UI9
 
 | | Atomic structure of the human Anaphase-Promoting Complex | | 分子名称: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | 著者 | Chang, L, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2015-03-27 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Atomic Structure of the Apc and its Mechanism of Protein Ubiquitination
Nature, 522, 2015
|
|
6XMG
 
 | | Cryo-EM structure of Cas12g ternary complex | | 分子名称: | CRISPR-Cas, RNA (130-MER), RNA (5'-R(P*UP*UP*AP*AP*UP*GP*CP*GP*GP*UP*AP*GP*UP*UP*UP*AP*UP*CP*AP*CP*AP*GP*UP*U)-3'), ... | | 著者 | Chang, L, Li, Z, Zhang, H. | | 登録日 | 2020-06-30 | | 公開日 | 2021-01-13 | | 最終更新日 | 2025-05-14 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Cryo-EM structure of the RNA-guided ribonuclease Cas12g.
Nat.Chem.Biol., 17, 2021
|
|
