1Z8M
 
 | |
9ATW
 
 | | Structure of biofilm-forming functional amyloid PSMa1 from Staphylococcus aureus | | Descriptor: | Phenol-soluble modulin alpha 1 peptide | | Authors: | Hansen, K.H, Byeon, C.H, Liu, Q, Drace, T, Boesen, T, Conway, J.F, Andreasen, M, Akbey, U. | | Deposit date: | 2024-02-27 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of biofilm-forming functional amyloid PSM alpha 1 from Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6N34
 
 | | Crystal structure of the BTB domain of Human NS1-BP | | Descriptor: | Influenza virus NS1A-binding protein | | Authors: | Zhang, K, Shang, G, Padavannil, A, Fontoura, B, Chook, Y.M. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6N3H
 
 | | Crystal structure of Kelch domain of the human NS1 binding protein | | Descriptor: | Influenza virus NS1A-binding protein | | Authors: | Zhang, K, Shang, G, Padavannil, A, Fontoura, B.M.A, Chook, Y.M. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7TBH
 
 | | cryo-EM structure of MBP-KIX-apoferritin complex with peptide 7 | | Descriptor: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed, LEU-SER-ARG-ARG-PRO-SEP-TYR-ARG-LYS-ILE-LEU-ASN-ASP-LEU-SER-SER-ASP-ALA-PRO | | Authors: | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
6XRZ
 
 | | The 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Descriptor: | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Authors: | Zhang, K, Zheludev, I, Hagey, R, Wu, M, Haslecker, R, Hou, Y, Kretsch, R, Pintilie, G, Rangan, R, Kladwang, W, Li, S, Pham, E, Souibgui, C, Baric, R, Sheahan, T, Souza, V, Glenn, J, Chiu, W, Das, R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-electron Microscopy and Exploratory Antisense Targeting of the 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome.
Biorxiv, 2020
|
|
7TB3
 
 | | cryo-EM structure of MBP-KIX-apoferritin | | Descriptor: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed | | Authors: | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
2OTR
 
 | |
8K58
 
 | | The cryo-EM map of close TIEA-TEC complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | TIEA inhibits Sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
8K5A
 
 | | The cryo-EM map of open TIEA-TEC complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | TIEA inhibits sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
6NYG
 
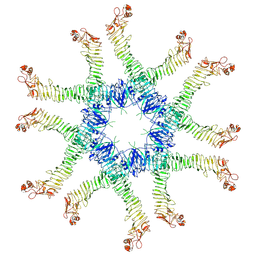 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2a (OA-2a) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYM
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2d (OA-2d) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYJ
 
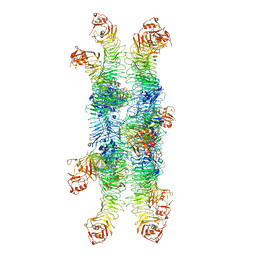 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2b (OA-2b) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYN
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2e (OA-2e) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYL
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2c (OA-2c) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYF
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 1 (OA-1) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3J1F
 
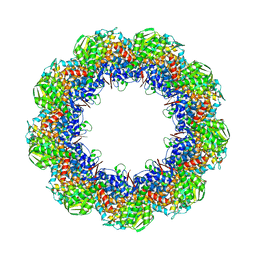 | | Cryo-EM structure of 9-fold symmetric rATcpn-beta in ATP-binding state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin beta subunit, MAGNESIUM ION | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
3J1E
 
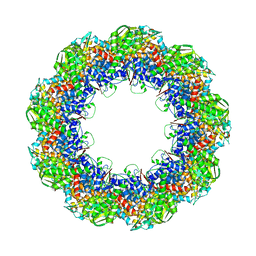 | | Cryo-EM structure of 9-fold symmetric rATcpn-beta in apo state | | Descriptor: | Chaperonin beta subunit | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
3J1B
 
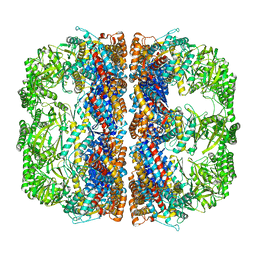 | | Cryo-EM structure of 8-fold symmetric rATcpn-alpha in apo state | | Descriptor: | Chaperonin alpha subunit | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
3J1C
 
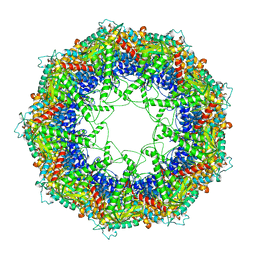 | | Cryo-EM structure of 9-fold symmetric rATcpn-alpha in apo state | | Descriptor: | Chaperonin alpha subunit | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
3EPU
 
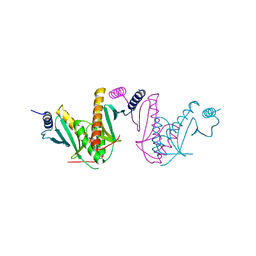 | | Crystal Structure of STM2138, a novel virulence chaperone in Salmonella | | Descriptor: | STM2138 Virulence Chaperone | | Authors: | Zhang, K, Andres, S.N, Hannemann, M, Coombes, B, Junop, M. | | Deposit date: | 2008-09-30 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis and quantitative proteomic
interactome of a novel virulence chaperone in Salmonella
To be Published
|
|
6NUD
 
 | | Small conformation of ssRNA-bound CRISPR_Csm complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms protein Csm2, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | Authors: | Zhang, K, Pintilie, G, Li, S, Zhu, Y, Chiu, W, Huang, Z. | | Deposit date: | 2019-01-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Coupling of ssRNA cleavage with DNase activity in type III-A CRISPR-Csm revealed by cryo-EM and biochemistry.
Cell Res., 29, 2019
|
|
6NUE
 
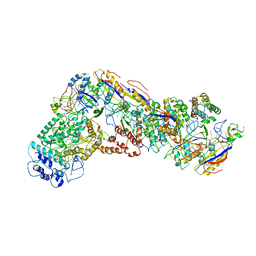 | | Small conformation of apo CRISPR_Csm complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms protein Csm2, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | Authors: | Zhang, K, Pintilie, G, Li, S, Zhu, Y, Chiu, W, Huang, Z. | | Deposit date: | 2019-01-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Coupling of ssRNA cleavage with DNase activity in type III-A CRISPR-Csm revealed by cryo-EM and biochemistry.
Cell Res., 29, 2019
|
|
7T32
 
 | | CryoEM structure of the adenosine 2A receptor-BRIL/Anti BRIL Fab complex with ZM241385 | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 Fusion Protein | | Authors: | Zhang, K.H, Wu, H, Hoppe, N, Manglik, A, Cheng, Y.F. | | Deposit date: | 2021-12-06 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Fusion protein strategies for cryo-EM study of G protein-coupled receptors.
Nat Commun, 13, 2022
|
|
5NVU
 
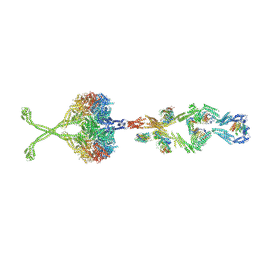 | | Full length human cytoplasmic dynein-1 in the phi-particle conformation | | Descriptor: | Dynein intermediate chain, Dynein light intermediate chain, Dynein motor domain, ... | | Authors: | Zhang, K, Foster, H.E, Carter, A.P. | | Deposit date: | 2017-05-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Cryo-EM Reveals How Human Cytoplasmic Dynein Is Auto-inhibited and Activated.
Cell, 169, 2017
|
|
